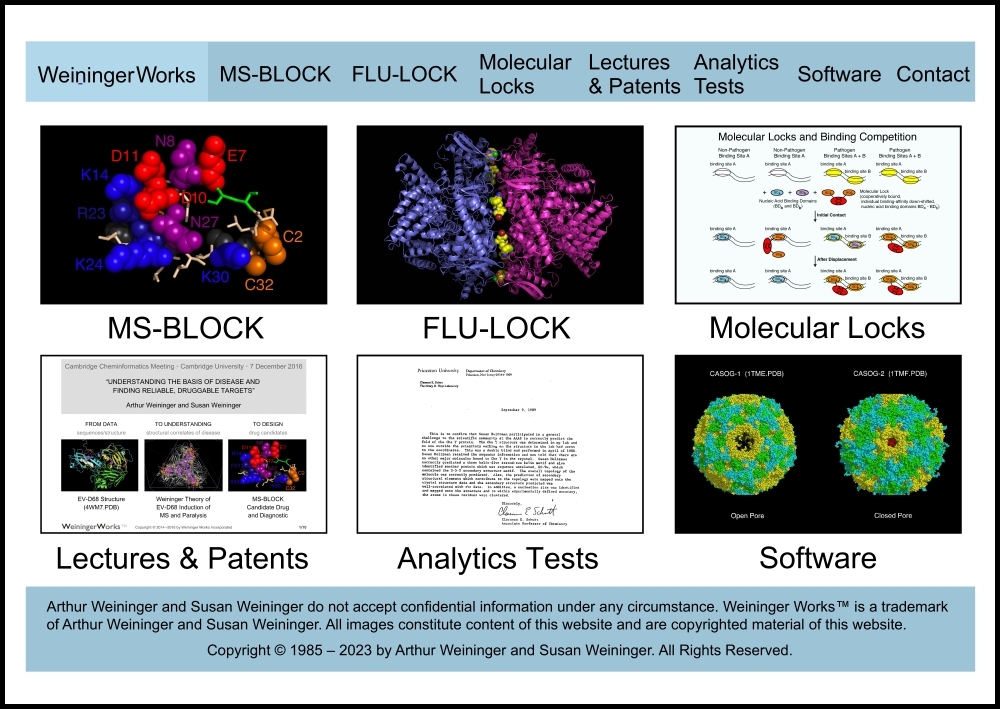
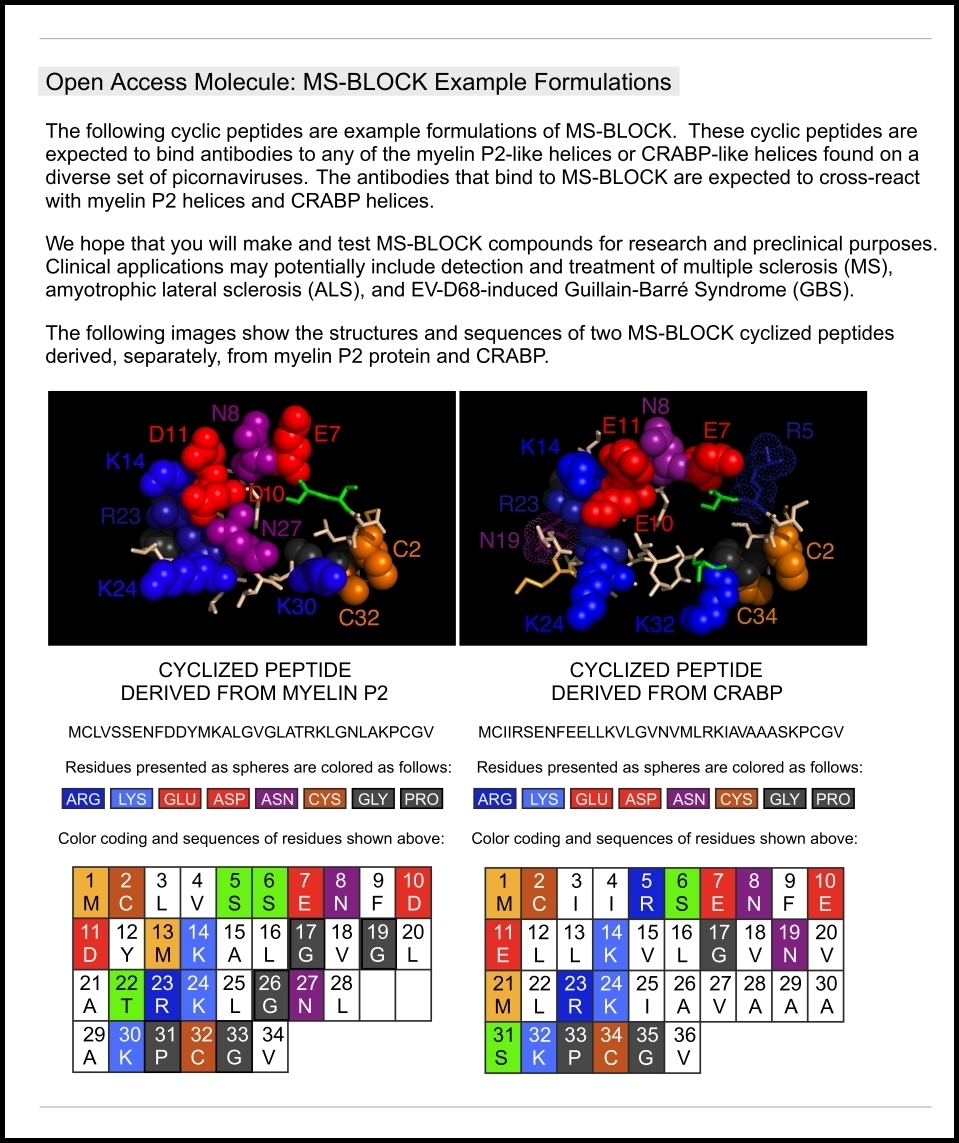
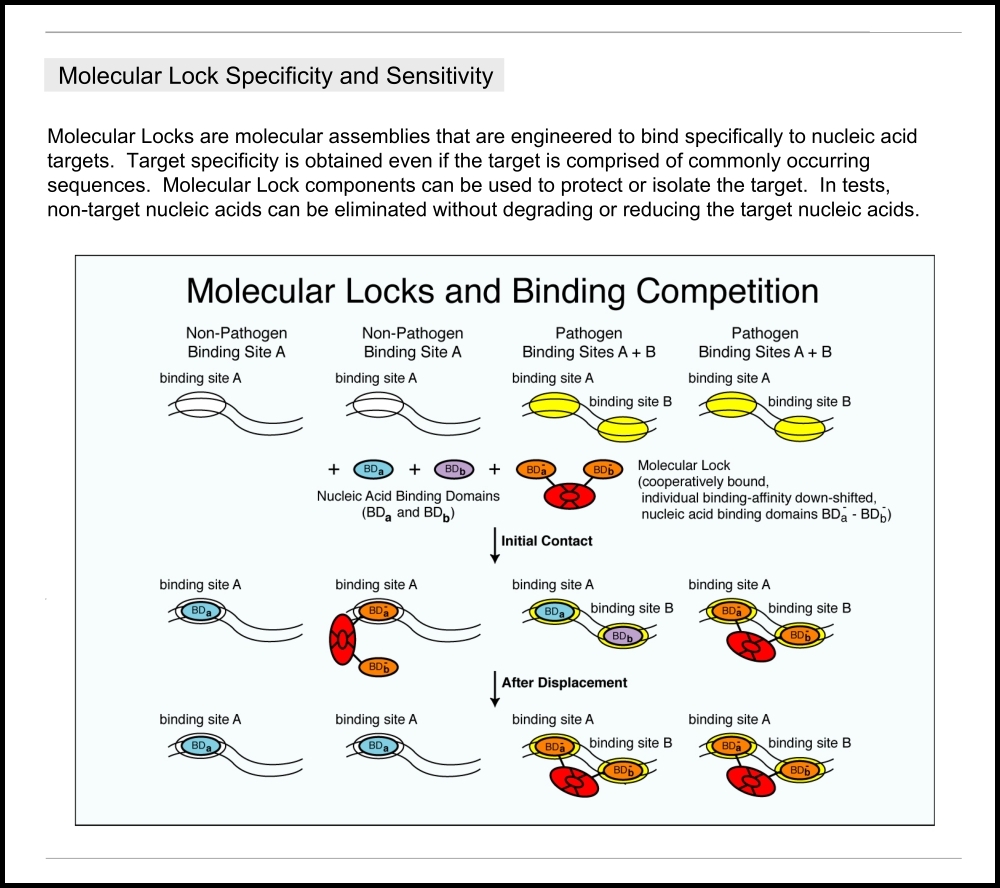
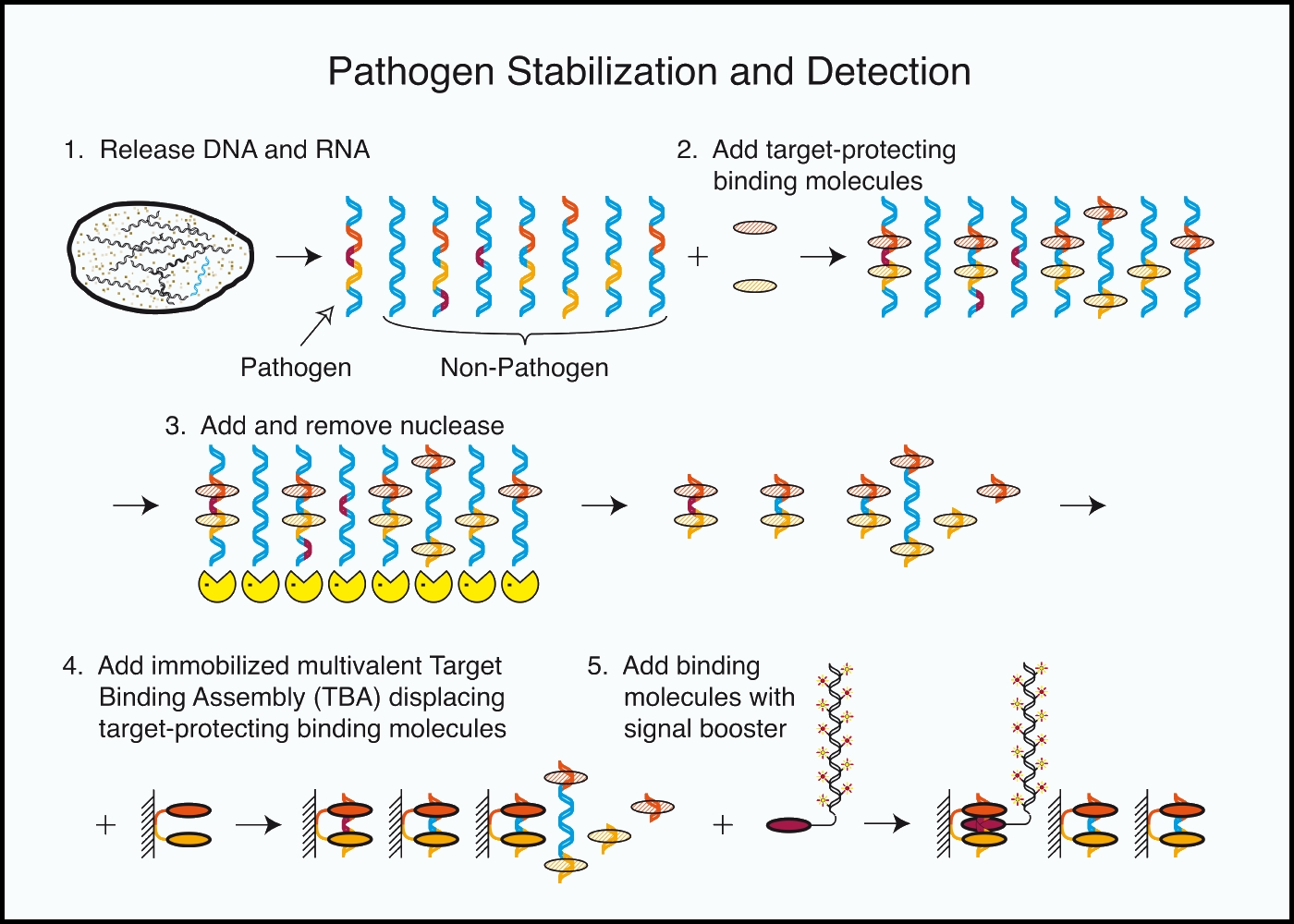
This “sitemap” page is a hierarchical list of weiningerworks.com.
All HTML links (internal and external) and downloads are listed.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Williams C, Thomas R, Pickersgill T, Lyons M, Lowe G, Stiff R, Moore C, Jones R, Howe R, Brunt H, Ashman A, Mason B. (2016)
Cluster of atypical adult Guillain-Barré syndrome temporally associated with neurological illness due to EV-D68 in children,
South Wales, United Kingdom, October 2015 to January 2016.
Euro Surveill. 21(4):pii=30119.
doi: 10.2807/1560-7917.ES.2016.21.4.30119
(EXTERNAL HTML REFERENCE)
|
|
|
|
|
|
Andrés C, Vila J, Creus-Costa A, Piñana M, González-Sánchez A, Esperalba J, Codina MG, Castillo C, Martín MC,
Fuentes F, Rubio S, García-Comuñas K, Vásquez-Mercado R, Saubi N, Rodrigo C, Pumarola T, Antón A (2022)
Enterovirus D68 in Hospitalized Children, Barcelona, Spain, 2014–2021.
Emerg Infect Dis. 2022 Jul;28(7): 1327-1331.
doi: 10.3201/eid2807.220264
(EXTERNAL HTML REFERENCE)
|
|
|
|
|
|
Harvala H, Broberg E, Benschop K, Berginc N, Ladhani S, Susi P,
Christiansen C, McKenna J, Allen D, Makiello P, McAllister G,
Carmen M, Zakikhany K, Dyrdak R, Nielsen X, Madsen T, Paul J,
Moore C, von Eije K, Piralla A, Carlier M, Vanoverschelde L,
Poelman R, Anton A, López-Labrador FX, Pellegrinelli L, Keeren K,
Maier M, Cassidy H, Derdas S, Savolainen-Kopra C, Diedrich S,
Nordbø S, Buesa J, Bailly JL, Baldanti F, MacAdam A, Mirand A,
Dudman S, Schuffenecker I, Kadambari K, Neyts J, Griffiths MJ,
Richter J, Margaretto C, Govind S, Morley U, Adams O, Krokstad S,
Dean J, Pons-Salort M, Prochazka B, Cabrerizo M, Majumdar M,
Nebbia G, Wiewel M, Cottrell S, Coyle P, Martin J, Moore C,
Midgley S, Horby P, Wolthers K, Simmonds P, Niesters H, Fischer TK. (2018)
Recommendations for enterovirus diagnostics and characterisation within and beyond Europe.
Journal of Clinical Virology 101 (2018) 11–17.
doi: 10.1016/j.jcv.2018.01.008
(EXTERNAL HTML REFERENCE)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
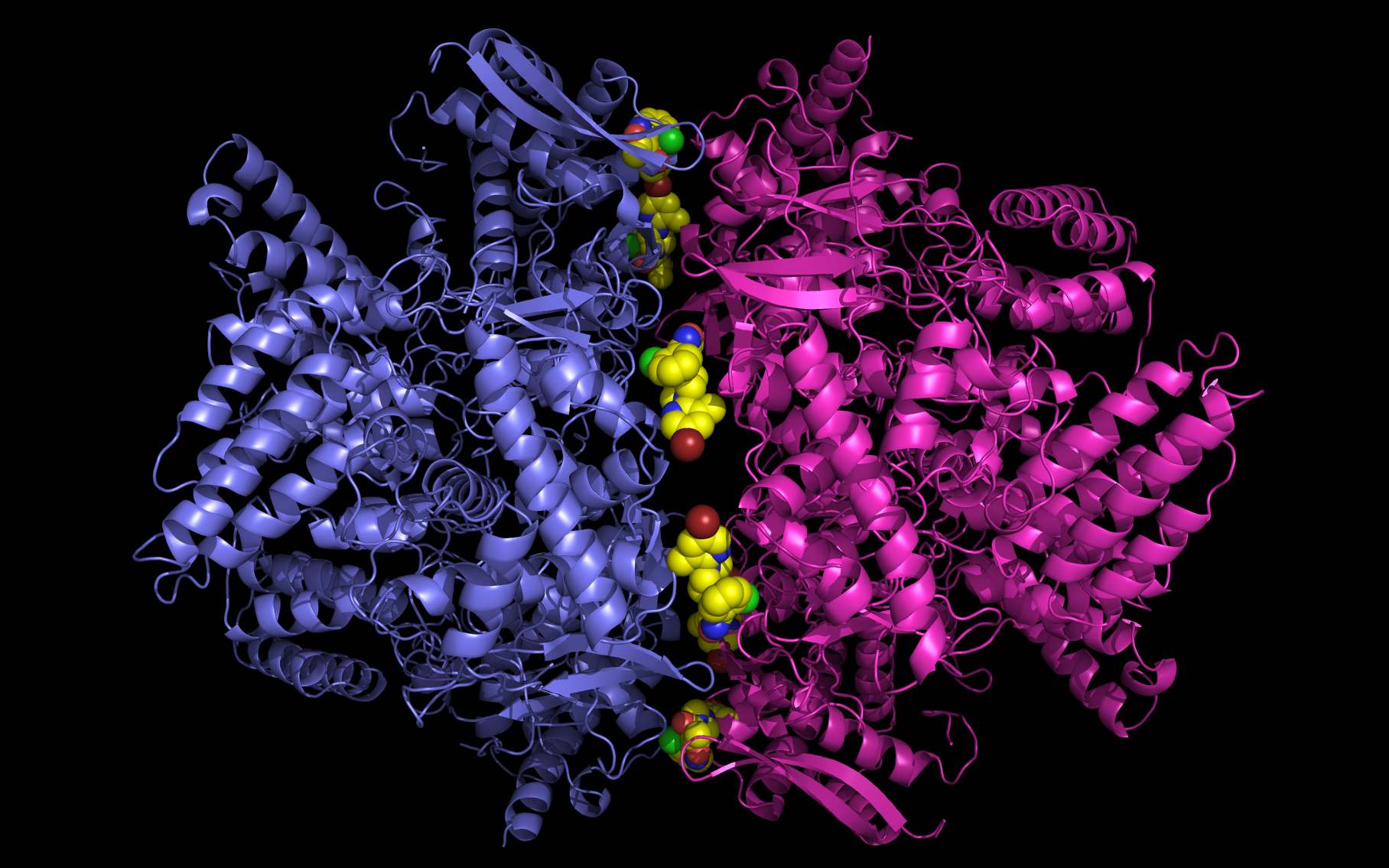
Li, Q., Sun, X., Li, Z., Liu, Y., Vavricka, C. J., Qi, J., & Gao, G. F. (2012).
Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus.
Proceedings of the National Academy of Sciences,
109(46), 18897-18902.
dx.doi.org/10.1073/pnas.1211037109
(EXTERNAL HTML REFERENCE)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
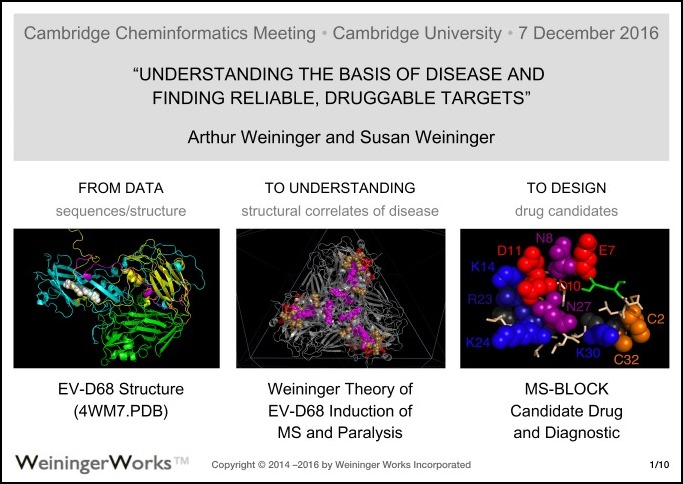
|
“Understanding the Basis of Disease and
Finding Reliable, Druggable Targets”
Arthur Weininger and Susan Weininger
Cambridge Cheminformatics Meeting
Cambridge University
7 December 2016
(PDF) of “slides“
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
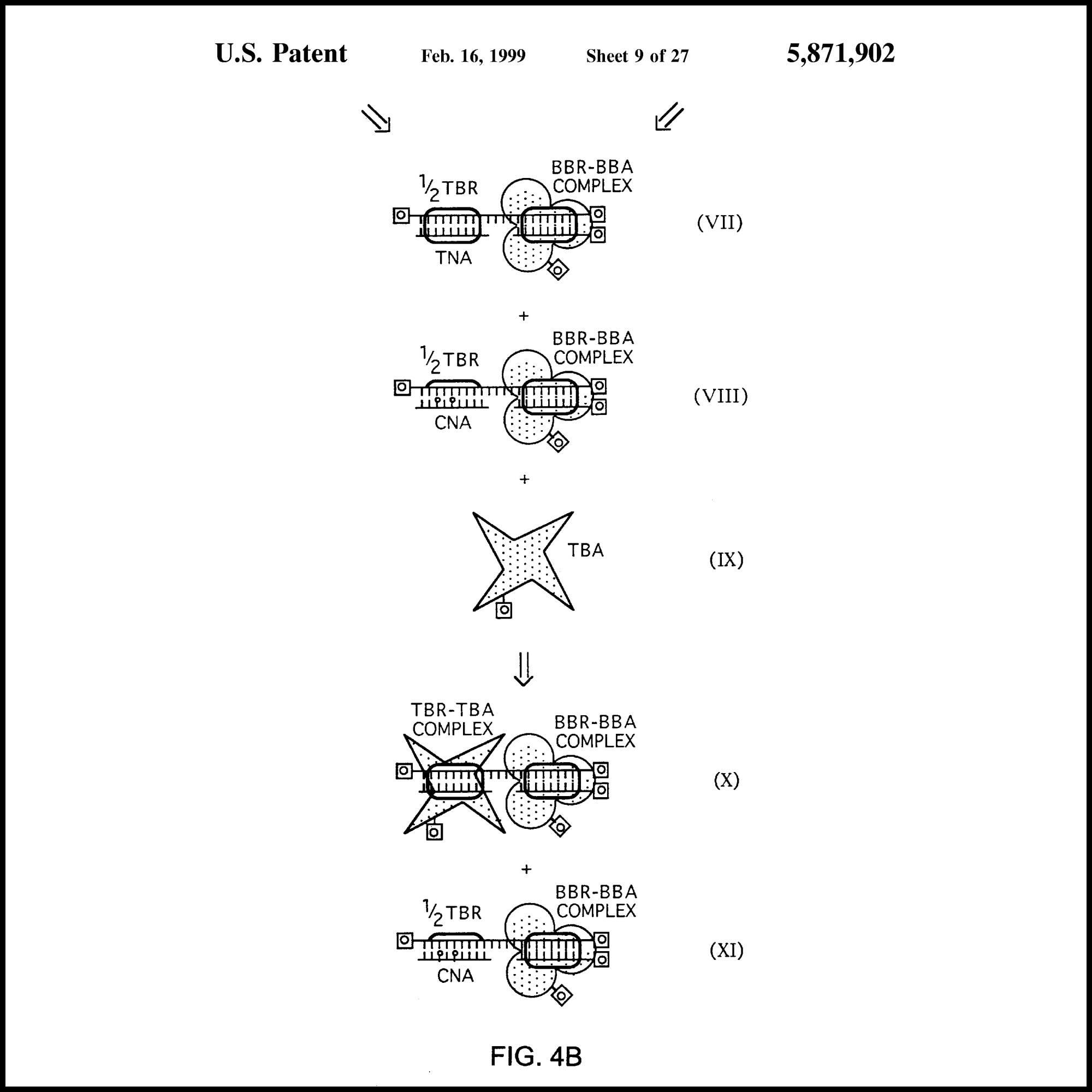
|
MS-BLOCK Patents
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|