Picornavirus_Superposition_Script
SCRIPT Overview
SCRIPT Related Downloads
SCRIPT Help Output
SCRIPT Execution Terminal Output
(WEB PAGE PDF)
SCRIPT Overview
The shell script “picornavirus_superposition_script.sh” creates PDB files showing the structural alignments described in the monograph titled “Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction” by Arthur Weininger and Susan Weininger. This shell script uses the Weininger Works’ unix command line programs “chainidPDB”, “combinePDB”, “stripPDB”, and “twwistPDB” to calculate molecular superpositions and combine multiple structures. This is a BASH (unix) shell script that should run on any of the Weininger Works’ supported operating systems/hardware.
SCRIPT Related Downloads
Monograph “Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction” by Arthur Weininger and Susan Weininger: (equivalent content)
- visit HTML pages
- download PDF File [24M]
- download PDF File [12M]
- download PDF File [6M]
- download PDF File [3M]
Picornavirus_Superposition_Scripts:
- picornavirus_superposition_script.sh
(complete script supporting '-c' ANSI/VT100 color and '-h' help options) - picornavirus_superposition_simple_script.sh
(simpler script with no options, which is more appropriate for editing)
Supporting Information PDB structure files:
- ICOS135.pdb
- TRI78.pdb
- CASOG_ONE_CAPSID_POINTS.pdb
- CASOG_TWO_CAPSID_POINTS.pdb
- PICORNAVIRUS_TILING_POINTS.pdb
PDB structure files (available from GenBank):
- 1AYM.pdb
- 1BBT.pdb
- 1BEV.pdb
- 1BL8.pdb
- 1CBS.pdb
- 1D4M.pdb
- 1EAH.pdb
- 1HXS.pdb
- 1OOP.pdb
- 1PIV.pdb
- 1TME.pdb
- 1TMF.pdb
- 1V9U.pdb
- 1W1X.pdb
- 1Z7S.pdb
- 2ABX.pdb
- 2WUT.pdb
- 3VBH.pdb
- 4CEW.pdb
- 4Q4Y.pdb
- 4WM7.pdb
Weininger Works’ software programs “chainidPDB”, “combinePDB”, “stripPDB”, and “twwistPDB”:
SCRIPT Help Output (“./picornavirus_superposition_script.sh -h” output)
NAME
picornavirus_superposition_script - a script to make specific superpositions
SYNOPSIS
picornavirus_superposition_script.sh [-c|-h]
DESCRIPTION
This shell script creates PDB files showing the structural alignments described
in the monograph titled “Common Features in Picornaviruses, Alpha-bungarotoxin,
Myelin P2, and CRABP Suggest Structural Bases for Multiple Sclerosis, Guillain-
Barré Syndrome, and Paralysis Induction” by Arthur Weininger and Susan Weininger.
The monograph is available on-line:
www.weiningerworks.com/picornavirus_monograph.html (HTML)
www.weiningerworks.com/picornavirus_monograph_12MB.pdf (PDF)
www.weiningerworks.com/picornavirus_monograph_6MB.pdf (PDF)
This shell script is available on-line:
www.weiningerworks.com/picornavirus_superposition_script.html (HTML)
www.weiningerworks.com/misc/picornavirus_superposition_script.sh (TEXT)
www.weiningerworks.com/misc/picornavirus_superposition_simple_script.sh (TEXT)
Input consists of PDB structure files (available from GenBank) and PDB files
representing geometric points (available from www.weiningerworks.com).
See the “STARTING DATA” section below for more information on input files.
Input files need to be placed in a subdirectory "“orig” that is located in
the directory containing the file “picornavirus_superposition_script.sh”.
Output consists of new PDB files that show either a monograph superposition
or a monograph figure. A subdirectory, named “new”, will be created in the
directory containing “picornavirus_superposition_script.sh”. Two new
directories, named “super” and “figs”, will be created in the “new” directory.
PDB files for each superposition in the monograph will be output to “new/super”.
PDB files for individual panels of monograph figures will output to “new/figs”;
these PDB files will be named by figure and panel.
This shell script displays progress by echoing running commands to “stdout”.
Any errors and warnings will be output to “stderr”. (“stdout” and “stderr”
are unix text streams that normally default to the terminal screen.)
This shell script is not interactive. This shell script does not require any
command line options. Option '-h' displays this help. Use the option '-c' if
you want to add ANSI/VT100 color to the terminal output of this shell script.
Color is used to delineate monograph references, file references, explanatory
text, unix commands, and error output. Color output is off by default.
This shell script uses the Weininger Works’ software programs “chainidPDB”,
“combinePDB”, “stripPDB”, and “twwistPDB” to calculate molecular superpositions
and combine multiple structures. These programs use the unix command line.
These programs are part of a software bundle supplied as a software installer.
This software installer may be requested from www.weiningerworks.com.
Licensed use of these software programs is subject to a license.
Agreement to this license is required by the installer.
We find our unix command line programs “chainidPDB”, “combinePDB”, “stripPDB”,
and “twwistPDB” to be simple and concise tools for performing superpositions.
While we used our software programs in this shell script, we appreciate that
there are many alternative means of calculating molecular superpositions and
of combining multiple structures (e.g., any molecular visualization program
having a pair-fitting function and that allows multiple structures to be read
in separately and then written out together.) We encourage use of more
familiar molecular analysis tools.
Note that this shell script does not necessarily produce structure files that,
when loaded into a molecular visualization program, will produce identical
images to those in our monograph. It does produce structure files that one can
use to produce identical images.
(i) Unless otherwise specified, our monograph images do not show hydrogen
atoms; water atoms, and other HETATMs.
(ii) Our monograph images often show a specific part of a protein structure;
many images hide parts of more complete structures to show a specific
structure.
(iii) Our monograph images show specific structures in specific colors and in
specific styles.
(iv) Our monograph images show specific structures from a specific perspective
(i.e., a specific orientation seen from a specific viewpoint).
This shell script is intended to create structural files that provide the
structural alignments specified in our monograph. Most molecular visualization
programs have the ability to hide specific atoms, set the color and appearance
of the structures being displayed, and change the displayed perspective.
“picornavirus_superposition_simple_script.sh” is an equivalent, but simpler,
shell script that is provided for modification purposes. This script either
can be downloaded from the www link above, or can be created by capturing the
terminal output from a picornavirus_superposition_script successfully run
without errors:
./picornavirus_superposition_script.sh > simple_script
The same amount of “disk space” is required for the execution of
“picornavirus_superposition_script.sh” and
“picornavirus_superposition_simple_script.sh”:
Initial Data ........................................... 12 MB
Generated PDB files showing monograph superpositions ... 127 MB
Generated PDB files showing individual figure panels ... 213 MB
---------------------------------------------------------------
Total required disk space .............................. 352 MB
The following is a list of monograph images and associated (super)structures.
MONOGRAPH MONOGRAPH MONOGRAPH
FIGURE FIGURE LOCATION TABLE(S) STRUCTURE: EXTERNAL_SOURCE
-------- --------------------- ----------- ------------------------------------
Figure 4 Column 1 Table 2 Monomers of: 1AYM, 1BBT, 1BEV, 1D4M,
1EAH, 1HXS, 1OOP, 1PIV,
1TME, 1TMF, 1V9U, 1Z7S,
3VBH, 4CEW, 4Q4Y, 4WM7
Figure 4 Columns 2 - 6, Row A Tables 3, 4 Trimers of: 1AYM, 1BBT, 1BEV, 1D4M,
1EAH, 1HXS, 1OOP, 1PIV,
1TME, 1TMF, 1V9U, 1Z7S,
3VBH, 4CEW, 4Q4Y, 4WM7
Figure 4 Columns 2 - 6, Row B Table 3 Trimer of: 1AYM
Figure 4 Columns 2 - 6, Row C Table 4 Trimer of: 1BBT
Figure 4 Columns 2 - 6, Row D Table 3 Trimer of: 1BEV
Figure 4 Columns 2 - 6, Row E Table 3 Trimer of: 1D4M
Figure 4 Columns 2 - 6, Row F Table 3 Trimer of: 1EAH
Figure 4 Columns 2 - 6, Row G Table 3 Trimer of: 1HXS
Figure 4 Columns 2 - 6, Row H Table 3 Trimer of: 1OOP
Figure 4 Columns 2 - 6, Row I Table 3 Trimer of: 1PIV
Figure 4 Columns 2 - 6, Row J Table 3 Trimer of: 1TME
Figure 4 Columns 2 - 6, Row K Table 4 Trimer of: 1TMF
Figure 4 Columns 2 - 6, Row L Table 3 Trimer of: 1V9U
Figure 4 Columns 2 - 6, Row M Table 3 Trimer of: 1Z7S
Figure 4 Columns 2 - 6, Row N Table 3 Trimer of: 3VBH
Figure 4 Columns 2 - 6, Row O Table 3 Trimer of: 4CEW
Figure 4 Columns 2 - 6, Row P Table 3 Trimer of: 4Q4Y
Figure 4 Columns 2 - 6, Row Q Table 3 Trimer of: 4WM7
Figure 5 Panels G, H, I Table 3 Multiple
Monomers of: 1TME
Figure 5 Panels M, N, O Table 4 Multiple
Monomers of: 1TMF
Figure 5 Panels P, S, R, U Table 3 Trimer of: 1TME
Figure 5 Panels Q, T, R, U Table 4 Trimer of: 1TMF
Figure 6 Panels A, D, G, J, Table 3 Multiple
C, F, I, L Trimers of: 1TME
Figure 6 Panels B, E, H, K, Table 4 Multiple
C, F, I, L Trimers of: 1TMF
Figure 7 Panels C, D Table 5 Trimers of: 2WUT, 1CBS
Figure 7 Panels E, F, G, H Tables 3, 6 Trimers of: 4WM7, 2ABX
Figure 8 Column 0 Table 5 Trimer of: 1CBS
Figure 8 Column 1 Table 5 Trimer of: 2WUT
Figure 8 Columns 2, 3 Table 4 Trimers of: 1BBT, 1TMF
Figure 8 Columns 4, 5, 6 Table 3 Trimers of: 1HXS, 1TME, 4WM7
Figure 9 Panels A, B Tables 3, 5 Trimers of: 2WUT, 2ABX
Figure 9 Panels C, D Table 4 Trimer of: 1BBT
Figure 9 Panels E, F Table 4 Trimer of: 1TMF
Figure 9 Panels G, H Table 3 Trimer of: 1TME
Figure 9 Panels I, J Table 3 Trimer of: 1HXS
Figure 9 Panels K, L Table 3 Trimer of: 4WM7
Figure 10 Panels A, B Table 5 Trimer of: 2ABX
Figure 10 Panels C - L Table 3 Trimer of: 4WM7
Figure 11 Panels A, B Monomer of: 1W1X
Figure 11 Panels C, D Monomer of: 4WM7
Figure 11 Panels E, F Table 3 Trimer of: 4WM7
Figure 12 Panels A, D, G, J, M Table 3 Trimer of: 1TME
C, F, I, L, O
Figure 12 Panels B, E, H, K, N Table 4 Trimer of: 1TMF
C, F, I, L, O
Figure 12 Panel P, R Table 3 Multiple
Trimers of: 1TME
Figure 12 Panel Q, R Table 3 Multiple
Trimers of: 1TMF
Figure 13 Panels A, C Table 3 Trimer of: 1TME
Figure 13 Panels B, C Table 4 Trimer of: 1TMF
Figure 13 Panel D Table 4 Monomer of: 1TME
Figure 13 Panel D Table 4 Monomer of: 1TMF
Figure 14 Column 1 Table 4 Trimer of: 1BBT
Figure 14 Column 2 Table 4 Trimer of: 1TMF
Figure 14 Column 3 Table 3 Trimer of: 1TME
Figure 14 Column 4 Table 3 Trimer of: 1HXS
Figure 14 Column 5 Table 3 Trimer of: 4WM7
Figure 15 Row A Table 5 Trimer of: 2WUT
Figure 15 Row B Table 4 Trimer of: 1BBT
Figure 15 Row C Table 4 Trimer of: 1TMF
Figure 15 Row D Table 3 Trimer of: 1TME
Figure 15 Row E Table 3 Trimer of: 4WM7
Figure 15 Row F Table 3 Trimer of: 1HXS
Figure 15 Panels G1, G2 (Tetramer) 1BL8
Figure 15 Panels G3, G4 Table 4 Trimer of: 1TMF
STARTING DATA
Supporting Information Files:
ICOS135.pdb
135-Angstrom-on-an-edge icosahedron with Zn atom vertices.
TRI78.pdb
Icosahedron edge points consisting of 78-Angstrom-on-an-edge equilateral
triangles with Zn atom vertices that are positioned on a virtual
135-Angstrom-on-an-edge icosahedron such that the 78-Angstrom-on-an-edge
triangle vertices are located on the icosahedron edges 45 (and 90) Angstroms
away from the icosahedron vertices.
CASOG_ONE_CAPSID_POINTS.pdb
Icosahedron face points consisting of scalene triangles with Zn atom vertices
positioned on faces of a virtual 135-Angstrom-on-an-edge icosahedron (as shown
in ICOS135.pdb). These scalene triangles are positioned within, and share one
vertex position with, 78-Angstrom-on-an-edge equilateral triangles (as shown
in TRI78.pdb).
CASOG_TWO_CAPSID_POINTS.pdb
Icosahedron face points consisting of 26-Angstrom-on-an-edge equilateral
triangles with Zn atom vertices that are positioned on virtual
78-Angstrom-on-an-edge equilateral triangles (as shown in TRI78.pdb) such
that each 26-Angstrom triangle is located at a corner of, and coplanar with,
a 78-Angstrom triangle, and furthermore that the 78-Angstrom triangles are
positioned on a virtual 135 Angstrom-on-an-edge icosahedron (as shown in
ICOS135.pdb) such that the 78-Angstrom vertices are located on the
icosahedron edges 45 (and 90) Angstroms from the icosahedron vertices.
PICORNAVIRUS_TILING_POINTS.pdb
Picornavirus capsid target superposition points all map to the first face of
a virtual 135 Angstrom-on-an-edge icosahedron (ICOS135). These superposition
points are grouped by function in separate chains. CASOG ONE tiling points
have the chain identifier “1”. CASOG TWO tiling points have the chain
identifier “2”. CASOG points 3, 6, and 9 are identical in both listed CASOG
sets. Trimer points for both cellular retinoic-acid-binding protein (1CBS)
and myelin P2 (2WUT) are identical and include three CASOG ONE points
(chain 1), three CASOG TWO points (chain 2), and three additional points
(points 23, 26, and 29). The CASOG points used for myelin P2/CRABP
superposition are listed again in the myelin P2/CRABP superposition points.
The superposition points for myelin P2/CRABP have the chain identifier “3”.
PDB files (available from GenBank, rename to match filenames below):
1AYM.pdb
1BBT.pdb
1BEV.pdb
1BL8.pdb
1CBS.pdb
1D4M.pdb
1EAH.pdb
1HXS.pdb
1OOP.pdb
1PIV.pdb
1TME.pdb
1TMF.pdb
1V9U.pdb
1W1X.pdb
1Z7S.pdb
2ABX.pdb
2WUT.pdb
3VBH.pdb
4CEW.pdb
4Q4Y.pdb
4WM7.pdb
RUNNING THIS UNIX SHELL SCRIPT
0. If not already installed, install the software programs
“chainidPDB”, “combinePDB”, “stripPDB”, and “twwistPDB”.
a. Request the software installer from www.weiningerworks.com:
https://www.weiningerworks.com/contact.html
b. Read and follow the installer information page:
https://www.weiningerworks.com/installerinfo.html
c. Read and follow the newly installed “README-program_installation.txt”.
This describes how to permanently set your unix environment variable 'PATH'
so that you can run installed Weininger Works programs from the unix command
line from any directory.
1. Download the file “picornavirus_superposition_script.sh” from:
https://www.weiningerworks.com/misc/picornavirus_superposition_script.sh
2. Place the file “picornavirus_superposition_script.sh” in a directory.
This directory will be the current directory when running this script.
a. You will need “write access” to this directory.
(You should have permission to write to any subdirectory of your home directory.)
b. You will need about 360 MB of available file space:
12 MB for original starting data, 347 MB for new files.
3. Create a new directory “orig” in the directory where “picornavirus_superposition_script.sh”
is located.
4. Download the “Starting Data” files into the “orig” directory.
There are links for downloading these starting files at:
https://www.weiningerworks.com/picornavirus_superposition_script.html#downloads
--> Rename structural PDB files to match the filenames in “STARTING DATA”, above.
5. Run “picornavirus_superposition_script.sh”.
a. Open a “Terminal” window.
b. Set the current directory of the terminal window to the directory of step (2).
In the Terminal window, type “cd <directory_specification>” and hit the
RETURN/ENTER key. (Substitute the actual directory specification for
“<directory_specification>”. To facilitate text insertion, the graphical
user interfaces (GUI) of most modern operating systems allow either:
the icon of the directory to be dragged and dropped into the Terminal
window or the copying and pasting of the directory path.)
c. Run this script.
On a command line, type “./picornavirus_superposition_script.sh -c” and hit
the RETURN/ENTER key.
(If the text output to the terminal is not readable, or you wish output
without color, type “./picornavirus_superposition_script.sh” and hit the
RETURN/ENTER key.)
d. If necessary, fix permissions of this shell script file to be executable.
If you get a message similar to “Permission denied”, set the shell script
file to be executable by typing “chmod +x picornavirus_superposition_script.sh”
and hit the RETURN/ENTER key. And then try step (5c) again.
6. Examine the newly created PDB files in the newly created subdirectory “new”.
SCRIPT Execution Terminal Output
(“./picornavirus_monograph_superposiion_script -c” output)
# Enter "./picornavirus_superposition_script.sh -h" for help and more information.
# Enter "./picornavirus_superposition_script.sh -c" for ANSI/VT100 color.
####################################################################################################
## ##
## Brief Summary: picornavirus_superposition_script.sh ##
## ##
## This shell script creates PDB files showing the structural alignments described in the ##
## monograph titled "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, ##
## and CRABP Suggest Structural Bases for Multiple Sclerosis, Guillain-Barré Syndrome, ##
## and Paralysis Induction" by Arthur Weininger and Susan Weininger. ##
## This shell script and the monograph are available at www.weiningerworks.com. ##
## ##
## This shell script uses the Weininger Works' software programs "chainidPDB", "combinePDB", ##
## "stripPDB", and "twwistPDB" to calculate molecular superpositions and combine multiple ##
## structures. You can request these programs from www.weiningerworks.com. ##
## ##
## Input consists of PDB structure files (available from GenBank) and PDB files ##
## representing geometric points (available from www.weiningerworks.com). ##
## Input location is a subdirectory of the current shell directory for this script, named 'orig'.##
## All new files generated by this script will be written to 'new', a script-created ##
## subdirectory of the current shell directory for this script, or to subdirectories of 'new'. ##
## ##
## For more script information, enter "./picornavirus_superposition_script.sh -h". ##
## ##
####################################################################################################
# New PDB files are made to show the molecular superpositions in the monograph.
# New subdirectories are made for new files.
#
# This script first makes PDB files for each superposition in the monograph; these files will
# be named by content and written out to 'super', a subdirectory of 'new'. Capsid tiles are
# generated for complete capsid models of 1TME and 1TMF. These PDB files are placed into new
# subdirectories of 'super' named '1TME_capsid_tiles' and '1TMF_capsid_tiles', respectively.
mkdir new;
mkdir new/super;
mkdir new/super/1TME_capsid_tiles;
mkdir new/super/1TMF_capsid_tiles;
# Strip HETATMs from PDBs.
#
# The majority of the figures in this monograph do not show HETATMs. Most commands in this
# superposition script use, directly or indirectly, copies of original PDB files that have
# had their HETATMs removed. This does not affect any of the superpositions. When there
# is a superposition or a figure that requires HETATMs, then the original PDB files are used.
# The following 'stripPDB' commands make copies, with no HETATMS, of the original PDB files.
# (Substitute the original PDB files if you wish to examine the superpositions with HETATMs.)
stripPDB --input=orig/1AYM.pdb --output=new/1AYMx.pdb --hetatms;
stripPDB --input=orig/1BBT.pdb --output=new/1BBTx.pdb --hetatms;
stripPDB --input=orig/1BEV.pdb --output=new/1BEVx.pdb --hetatms;
stripPDB --input=orig/1CBS.pdb --output=new/1CBSx.pdb --hetatms;
stripPDB --input=orig/1D4M.pdb --output=new/1D4Mx.pdb --hetatms;
stripPDB --input=orig/1EAH.pdb --output=new/1EAHx.pdb --hetatms;
stripPDB --input=orig/1HXS.pdb --output=new/1HXSx.pdb --hetatms;
stripPDB --input=orig/1OOP.pdb --output=new/1OOPx.pdb --hetatms;
stripPDB --input=orig/1PIV.pdb --output=new/1PIVx.pdb --hetatms --remove="0";
stripPDB --input=orig/1TME.pdb --output=new/1TMEx.pdb --hetatms;
stripPDB --input=orig/1TMF.pdb --output=new/1TMFx.pdb --hetatms;
stripPDB --input=orig/1V9U.pdb --output=new/1V9Ux.pdb --hetatms --remove="5";
stripPDB --input=orig/1W1X.pdb --output=new/1W1Xx.pdb --hetatms;
stripPDB --input=orig/1Z7S.pdb --output=new/1Z7Sx.pdb --hetatms;
stripPDB --input=orig/2ABX.pdb --output=new/2ABXx.pdb --hetatms;
stripPDB --input=orig/2WUT.pdb --output=new/2WUTx.pdb --hetatms;
stripPDB --input=orig/3VBH.pdb --output=new/3VBHx.pdb --hetatms;
stripPDB --input=orig/4CEW.pdb --output=new/4CEWx.pdb --hetatms;
stripPDB --input=orig/4Q4Y.pdb --output=new/4Q4Yx.pdb --hetatms;
stripPDB --input=orig/4WM7.pdb --output=new/4WM7x.pdb --hetatms;
# "VP1234" PDBs are altered to have consistent chain identifiers.
#
# The VP1234 picornavirus PDBs examined in the monograph either have chain identifier labels
# of 'A', 'B', 'C', 'D' or of '1', '2', '3', '4'. For consistency and ease of manipulation,
# copies are made of the PDB files in order to have similarly named chain identifiers. These
# copies are identical to the original PDB files with the exception that the chain identifiers
# '1', '2', '3', and '4' are respectively changed to 'A', 'B', 'C', and 'D'.
chainidPDB --input=new/1AYMx.pdb --output=new/1AYMxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1AYMxx.pdb new/1AYMx.pdb;
chainidPDB --input=new/1BBTx.pdb --output=new/1BBTxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1BBTxx.pdb new/1BBTx.pdb;
chainidPDB --input=new/1BEVx.pdb --output=new/1BEVxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1BEVxx.pdb new/1BEVx.pdb;
chainidPDB --input=new/1CBSx.pdb --output=new/1CBSxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1CBSxx.pdb new/1CBSx.pdb;
chainidPDB --input=new/1D4Mx.pdb --output=new/1D4Mxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1D4Mxx.pdb new/1D4Mx.pdb;
chainidPDB --input=new/1EAHx.pdb --output=new/1EAHxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1EAHxx.pdb new/1EAHx.pdb;
chainidPDB --input=new/1HXSx.pdb --output=new/1HXSxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1HXSxx.pdb new/1HXSx.pdb;
chainidPDB --input=new/1PIVx.pdb --output=new/1PIVxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1PIVxx.pdb new/1PIVx.pdb;
chainidPDB --input=new/1TMEx.pdb --output=new/1TMExx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1TMExx.pdb new/1TMEx.pdb;
chainidPDB --input=new/1TMFx.pdb --output=new/1TMFxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1TMFxx.pdb new/1TMFx.pdb;
chainidPDB --input=new/1V9Ux.pdb --output=new/1V9Uxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1V9Uxx.pdb new/1V9Ux.pdb;
chainidPDB --input=new/1W1Xx.pdb --output=new/1W1Xxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1W1Xxx.pdb new/1W1Xx.pdb;
chainidPDB --input=new/1Z7Sx.pdb --output=new/1Z7Sxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/1Z7Sxx.pdb new/1Z7Sx.pdb;
chainidPDB --input=new/4Q4Yx.pdb --output=new/4Q4Yxx.pdb --from="1234" --to="ABCD" --nopositionchange;
mv new/4Q4Yxx.pdb new/4Q4Yx.pdb;
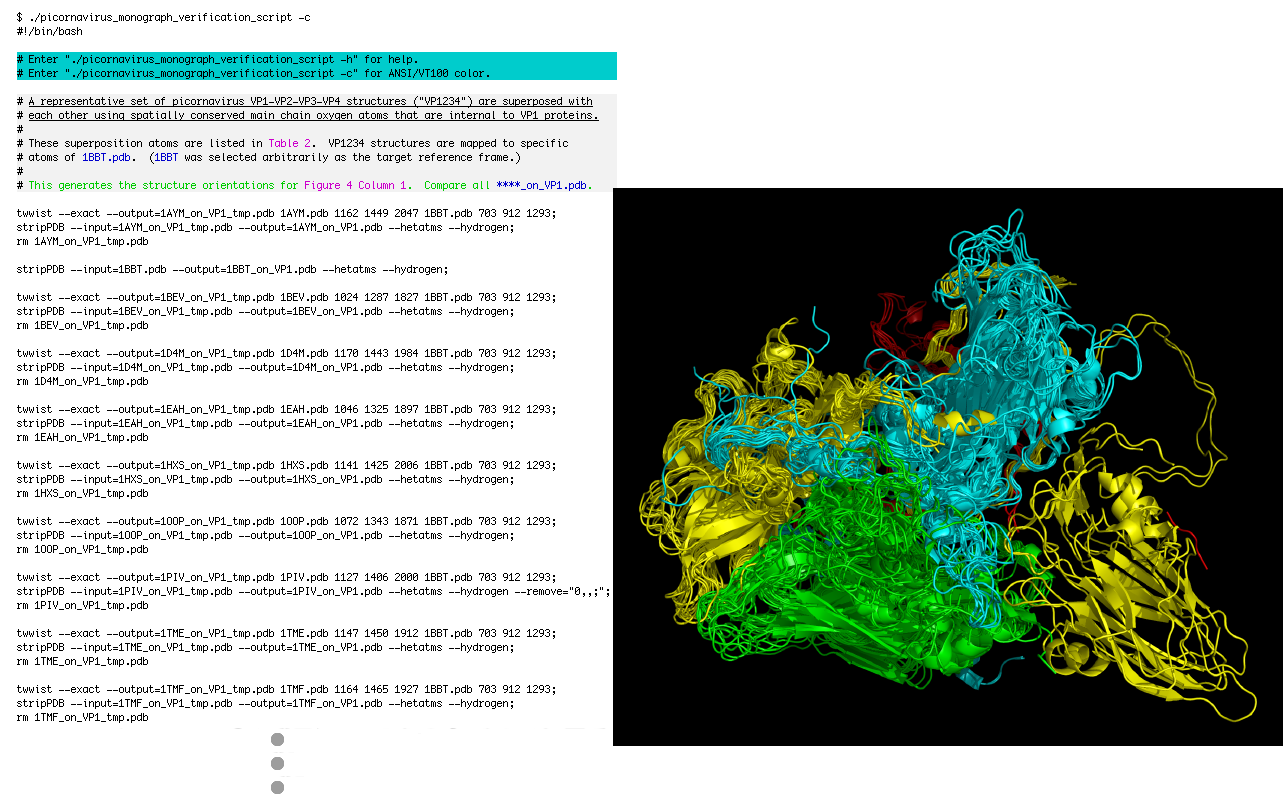
# A representative set of picornavirus VP1-VP2-VP3-VP4 structures ("VP1234") are superposed with
# each other using spatially conserved main chain oxygen atoms that are internal to VP1 proteins.
#
# These superposition atoms are listed in Table 2. VP1234 structures are mapped to specific
# atoms of 1BBT.pdb. (1BBT was selected arbitrarily as the target reference frame.)
#
# This generates the structure orientations for Figure 4 Column 1. Compare all ****_on_VP1.pdb.
twwistPDB --exact --output=new/super/1AYM_on_VP1.pdb new/1AYMx.pdb 1162 1449 2047 orig/1BBT.pdb 703 912 1293;
cp new/1BBTx.pdb new/super/1BBT_on_VP1.pdb;
twwistPDB --exact --output=new/super/1BEV_on_VP1.pdb new/1BEVx.pdb 1024 1287 1827 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1D4M_on_VP1.pdb new/1D4Mx.pdb 1170 1443 1984 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1EAH_on_VP1.pdb new/1EAHx.pdb 1046 1325 1897 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1HXS_on_VP1.pdb new/1HXSx.pdb 1141 1425 2006 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1OOP_on_VP1.pdb new/1OOPx.pdb 1072 1343 1871 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1PIV_on_VP1.pdb new/1PIVx.pdb 1127 1406 2000 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1TME_on_VP1.pdb new/1TMEx.pdb 1147 1450 1912 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1TMF_on_VP1.pdb new/1TMFx.pdb 1164 1465 1927 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1V9U_on_VP1.pdb new/1V9Ux.pdb 1047 1332 1857 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/1Z7S_on_VP1.pdb new/1Z7Sx.pdb 1107 1393 1980 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/3VBH_on_VP1.pdb new/3VBHx.pdb 1188 1454 2032 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/4CEW_on_VP1.pdb new/4CEWx.pdb 1188 1454 2030 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/4Q4Y_on_VP1.pdb new/4Q4Yx.pdb 1114 1396 1993 orig/1BBT.pdb 703 912 1293;
twwistPDB --exact --output=new/super/4WM7_on_VP1.pdb new/4WM7x.pdb 1079 1357 1850 orig/1BBT.pdb 703 912 1293;
# Construction of CASOG ONE and CASOG TWO model capsid tiles
#
# Three VP1234 assemblies are directly positioned onto an icosahedron face to construct a VP1234
# trimer ("3xVP1234 capsid tile"). Superpositions of specific VP1, VP2, and VP3 proline main chain
# oxygen atoms (listed in Tables 3 and 4) onto specific icosahedron face points (provided in
# supporting information files CASOG_ONE_CAPSID_POINTS.pdb and CASOG_TWO_CAPSID_POINTS.pdb)
# place individual VP1234 assemblies.
#
# The mapping order of CASOG ONE and CASOG TWO is straight: the atoms of VP1, VP2, and VP3 map to
# capsid superposition points (1, 2, 3), (4, 5, 6), etc.
#
# twwistPDB commands that map VP1234 assemblies to capsid face points in CASOG ONE orientation
# normally have the following format:
#
# twwistPDB initial_VP1234.pdb VP1 VP2 VP3 CASOG_ONE_CAPSID_POINTS.pdb N N+1 N+2 > VP1234_on_capsid.pdb
#
# The three commented twwistPDB commands, immediately below, are an example of normal twwistPDB commands.
#
# twwistPDB 1AYM.pdb 1162 3305 5375 CASOG_ONE_CAPSID_POINTS.pdb 1 2 3 > 1AYM_tile_IN_CASOG_ONE_1a.pdb;
# twwistPDB 1AYM.pdb 1162 3305 5375 CASOG_ONE_CAPSID_POINTS.pdb 4 5 6 > 1AYM_tile_IN_CASOG_ONE_1b.pdb;
# twwistPDB 1AYM.pdb 1162 3305 5375 CASOG_ONE_CAPSID_POINTS.pdb 7 8 9 > 1AYM_tile_IN_CASOG_ONE_1c.pdb;
#
# These three twwistPDB commands place three VP1234 assemblies on the first capsid icosahedron face.
# "combinePDB" is used to create a single capsid tiling piece from these reoriented VP1234 assemblies.
#
# The actual commands used, just below, are special in that they prioritize VP3 atom placement
# instead of minimizing the sum of all atom placement error. The order of the atoms listed in
# the twwistPDB commands below starts with the VP3 atom, the "--exact ('-e')" option is used,
# and the "--minimize ('-m')" option is not used. (This combination of options specifies that
# the first atoms will be exactly superposed, and that any existing differences in distances
# between the pairs of specified atoms in the two different molecules will not be minimized.
# The VP3 atom is the pivot point between CASOG ONE and CASOG TWO orientations.)
# The following format is used:
#
# twwistPDB --exact initVP1234.pdb VP3 VP1 VP2 CASOG_ONE_CAPSID_POINTS.pdb N+2 N N+1 > capsidVP1234.pdb
#
# This generates the structures for Figure 4 Column 2 Panels A2 - Q2. See ****_tile_1.pdb.
# The individal VP trimers shown in Figure 4 Columns 3 - 6 are substructures of these files.
#
# This generates the structures for Figure 5 Panels G - I and Panels M - U.
# See 1TME_tile_1a.pdb, 1TME_tile_1b.pdb, 1TME_tile_1c.pdb, 1TME_tile_1.pdb,
# 1TMF_tile_1a.pdb, 1TMF_tile_1b.pdb, 1TMF_tile_1c.pdb, 1TMF_tile_1.pdb.
#
# This generates the structures for Figure 7 Panels F, G, and H, Figure 10 Panels C - J, and
# Figure 11 Panels E and F. The VP1 trimers in these panels are substructures of 4WM7_tile_1.pdb.
#
# This generates the structures for Figure 8 Columns 2 - 6 and Figure 9 Panels C - L.
# See 1BBT_tile_1.pdb, 1TMF_tile_1.pdb, 1TME_tile_1.pdb, 1HXS_tile_1.pdb, and 4WM7_tile_1.pdb.
# The individal VP trimers shown in these images are substructures of these files.
#
# This generates the structures for Figure 12 Panels A - O and all the panels of Figure 13.
# See 1TME_tile_1.pdb and 1TMF_tile_1.pdb.
# The individal VP trimers shown in these images are substructures of these files.
#
# This generates the structures for all the columns of Figure 14.
# See 1BBT_tile_1.pdb, 1TMF_tile_1.pdb, 1TME_tile_1.pdb, 1HXS_tile_1.pdb, and 4WM7_tile_1.pdb.
# The individal VP trimers shown in these images are substructures of these files.
#
# This generates the structures for Figure 15 Rows B - F (all panels) and Figure 15 Panels G3 and G4.
# See 1BBT_tile_1.pdb, 1TMF_tile_1.pdb, 1TME_tile_1.pdb, 4WM7_tile_1.pdb, and 1HXS_tile_1.pdb.
# The individal VP trimers shown in these images are substructures of these files.
twwistPDB --exact new/1AYMx.pdb 5375 1162 3305 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1AYM_tile_1a.pdb;
twwistPDB --exact new/1AYMx.pdb 5375 1162 3305 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1AYM_tile_1b.pdb;
twwistPDB --exact new/1AYMx.pdb 5375 1162 3305 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1AYM_tile_1c.pdb;
combinePDB --input1=new/super/1AYM_tile_1a.pdb --input2=new/super/1AYM_tile_1b.pdb --output=new/super/1AYM_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1AYM_tile_1ab.pdb --input2=new/super/1AYM_tile_1c.pdb --output=new/super/1AYM_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1AYM_tile_1a.pdb;
rm new/super/1AYM_tile_1b.pdb;
rm new/super/1AYM_tile_1c.pdb;
rm new/super/1AYM_tile_1ab.pdb;
twwistPDB --exact new/1BBTx.pdb 4083 703 2375 orig/CASOG_TWO_CAPSID_POINTS.pdb 3 1 2 > new/super/1BBT_tile_1a.pdb;
twwistPDB --exact new/1BBTx.pdb 4083 703 2375 orig/CASOG_TWO_CAPSID_POINTS.pdb 6 4 5 > new/super/1BBT_tile_1b.pdb;
twwistPDB --exact new/1BBTx.pdb 4083 703 2375 orig/CASOG_TWO_CAPSID_POINTS.pdb 9 7 8 > new/super/1BBT_tile_1c.pdb;
combinePDB --input1=new/super/1BBT_tile_1a.pdb --input2=new/super/1BBT_tile_1b.pdb --output=new/super/1BBT_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1BBT_tile_1ab.pdb --input2=new/super/1BBT_tile_1c.pdb --output=new/super/1BBT_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1BBT_tile_1a.pdb;
rm new/super/1BBT_tile_1b.pdb;
rm new/super/1BBT_tile_1c.pdb;
rm new/super/1BBT_tile_1ab.pdb;
twwistPDB --exact new/1BEVx.pdb 5009 1024 3048 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1BEV_tile_1a.pdb;
twwistPDB --exact new/1BEVx.pdb 5009 1024 3048 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1BEV_tile_1b.pdb;
twwistPDB --exact new/1BEVx.pdb 5009 1024 3048 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1BEV_tile_1c.pdb;
combinePDB --input1=new/super/1BEV_tile_1a.pdb --input2=new/super/1BEV_tile_1b.pdb --output=new/super/1BEV_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1BEV_tile_1ab.pdb --input2=new/super/1BEV_tile_1c.pdb --output=new/super/1BEV_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1BEV_tile_1a.pdb;
rm new/super/1BEV_tile_1b.pdb;
rm new/super/1BEV_tile_1c.pdb;
rm new/super/1BEV_tile_1ab.pdb;
twwistPDB --exact new/1D4Mx.pdb 5266 1170 3207 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1D4M_tile_1a.pdb;
twwistPDB --exact new/1D4Mx.pdb 5266 1170 3207 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1D4M_tile_1b.pdb;
twwistPDB --exact new/1D4Mx.pdb 5266 1170 3207 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1D4M_tile_1c.pdb;
combinePDB --input1=new/super/1D4M_tile_1a.pdb --input2=new/super/1D4M_tile_1b.pdb --output=new/super/1D4M_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1D4M_tile_1ab.pdb --input2=new/super/1D4M_tile_1c.pdb --output=new/super/1D4M_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1D4M_tile_1a.pdb;
rm new/super/1D4M_tile_1b.pdb;
rm new/super/1D4M_tile_1c.pdb;
rm new/super/1D4M_tile_1ab.pdb;
twwistPDB --exact new/1EAHx.pdb 5220 1046 3077 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1EAH_tile_1a.pdb;
twwistPDB --exact new/1EAHx.pdb 5220 1046 3077 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1EAH_tile_1b.pdb;
twwistPDB --exact new/1EAHx.pdb 5220 1046 3077 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1EAH_tile_1c.pdb;
combinePDB --input1=new/super/1EAH_tile_1a.pdb --input2=new/super/1EAH_tile_1b.pdb --output=new/super/1EAH_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1EAH_tile_1ab.pdb --input2=new/super/1EAH_tile_1c.pdb --output=new/super/1EAH_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1EAH_tile_1a.pdb;
rm new/super/1EAH_tile_1b.pdb;
rm new/super/1EAH_tile_1c.pdb;
rm new/super/1EAH_tile_1ab.pdb;
twwistPDB --exact new/1HXSx.pdb 5376 1141 3227 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1HXS_tile_1a.pdb;
twwistPDB --exact new/1HXSx.pdb 5376 1141 3227 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1HXS_tile_1b.pdb;
twwistPDB --exact new/1HXSx.pdb 5376 1141 3227 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1HXS_tile_1c.pdb;
combinePDB --input1=new/super/1HXS_tile_1a.pdb --input2=new/super/1HXS_tile_1b.pdb --output=new/super/1HXS_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1HXS_tile_1ab.pdb --input2=new/super/1HXS_tile_1c.pdb --output=new/super/1HXS_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1HXS_tile_1a.pdb;
rm new/super/1HXS_tile_1b.pdb;
rm new/super/1HXS_tile_1c.pdb;
rm new/super/1HXS_tile_1ab.pdb;
twwistPDB --exact new/1OOPx.pdb 5106 1072 3081 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1OOP_tile_1a.pdb;
twwistPDB --exact new/1OOPx.pdb 5106 1072 3081 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1OOP_tile_1b.pdb;
twwistPDB --exact new/1OOPx.pdb 5106 1072 3081 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1OOP_tile_1c.pdb;
combinePDB --input1=new/super/1OOP_tile_1a.pdb --input2=new/super/1OOP_tile_1b.pdb --output=new/super/1OOP_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1OOP_tile_1ab.pdb --input2=new/super/1OOP_tile_1c.pdb --output=new/super/1OOP_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1OOP_tile_1a.pdb;
rm new/super/1OOP_tile_1b.pdb;
rm new/super/1OOP_tile_1c.pdb;
rm new/super/1OOP_tile_1ab.pdb;
twwistPDB --exact new/1PIVx.pdb 5378 1127 3223 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1PIV_tile_1a.pdb;
twwistPDB --exact new/1PIVx.pdb 5378 1127 3223 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1PIV_tile_1b.pdb;
twwistPDB --exact new/1PIVx.pdb 5378 1127 3223 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1PIV_tile_1c.pdb;
combinePDB --input1=new/super/1PIV_tile_1a.pdb --input2=new/super/1PIV_tile_1b.pdb --output=new/super/1PIV_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1PIV_tile_1ab.pdb --input2=new/super/1PIV_tile_1c.pdb --output=new/super/1PIV_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1PIV_tile_1a.pdb;
rm new/super/1PIV_tile_1b.pdb;
rm new/super/1PIV_tile_1c.pdb;
rm new/super/1PIV_tile_1ab.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1TME_tile_1a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1TME_tile_1b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1TME_tile_1c.pdb;
combinePDB --input1=new/super/1TME_tile_1a.pdb --input2=new/super/1TME_tile_1b.pdb --output=new/super/1TME_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1TME_tile_1ab.pdb --input2=new/super/1TME_tile_1c.pdb --output=new/super/1TME_tile_1.pdb --chainid="ABCDEFGHIJKL";
#rm new/super/1TME_tile_1a.pdb;
#rm new/super/1TME_tile_1b.pdb;
#rm new/super/1TME_tile_1c.pdb;
rm new/super/1TME_tile_1ab.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 3 1 2 > new/super/1TMF_tile_1a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 6 4 5 > new/super/1TMF_tile_1b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 9 7 8 > new/super/1TMF_tile_1c.pdb;
combinePDB --input1=new/super/1TMF_tile_1a.pdb --input2=new/super/1TMF_tile_1b.pdb --output=new/super/1TMF_tile_1ab.pdb --chainid="ABCDEFGH" --map=new/super/1TMF_tile_ab.map;
combinePDB --input1=new/super/1TMF_tile_1ab.pdb --input2=new/super/1TMF_tile_1c.pdb --output=new/super/1TMF_tile_1.pdb --chainid="ABCDEFGHIJKL" --map=new/super/1TMF_tile_abc.map;
rm new/super/1TMF_tile_1a.pdb;
rm new/super/1TMF_tile_1b.pdb;
rm new/super/1TMF_tile_1c.pdb;
rm new/super/1TMF_tile_1ab.pdb;
twwistPDB --exact new/1V9Ux.pdb 5161 1047 3077 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1V9U_tile_1a.pdb;
twwistPDB --exact new/1V9Ux.pdb 5161 1047 3077 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1V9U_tile_1b.pdb;
twwistPDB --exact new/1V9Ux.pdb 5161 1047 3077 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1V9U_tile_1c.pdb;
combinePDB --input1=new/super/1V9U_tile_1a.pdb --input2=new/super/1V9U_tile_1b.pdb --output=new/super/1V9U_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1V9U_tile_1ab.pdb --input2=new/super/1V9U_tile_1c.pdb --output=new/super/1V9U_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1V9U_tile_1a.pdb;
rm new/super/1V9U_tile_1b.pdb;
rm new/super/1V9U_tile_1c.pdb;
rm new/super/1V9U_tile_1ab.pdb;
twwistPDB --exact new/1Z7Sx.pdb 5338 1107 3204 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1Z7S_tile_1a.pdb;
twwistPDB --exact new/1Z7Sx.pdb 5338 1107 3204 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1Z7S_tile_1b.pdb;
twwistPDB --exact new/1Z7Sx.pdb 5338 1107 3204 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1Z7S_tile_1c.pdb;
combinePDB --input1=new/super/1Z7S_tile_1a.pdb --input2=new/super/1Z7S_tile_1b.pdb --output=new/super/1Z7S_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/1Z7S_tile_1ab.pdb --input2=new/super/1Z7S_tile_1c.pdb --output=new/super/1Z7S_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/1Z7S_tile_1a.pdb;
rm new/super/1Z7S_tile_1b.pdb;
rm new/super/1Z7S_tile_1c.pdb;
rm new/super/1Z7S_tile_1ab.pdb;
twwistPDB --exact new/3VBHx.pdb 5243 1188 3240 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/3VBH_tile_1a.pdb;
twwistPDB --exact new/3VBHx.pdb 5243 1188 3240 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/3VBH_tile_1b.pdb;
twwistPDB --exact new/3VBHx.pdb 5243 1188 3240 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/3VBH_tile_1c.pdb;
combinePDB --input1=new/super/3VBH_tile_1a.pdb --input2=new/super/3VBH_tile_1b.pdb --output=new/super/3VBH_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/3VBH_tile_1ab.pdb --input2=new/super/3VBH_tile_1c.pdb --output=new/super/3VBH_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/3VBH_tile_1a.pdb;
rm new/super/3VBH_tile_1b.pdb;
rm new/super/3VBH_tile_1c.pdb;
rm new/super/3VBH_tile_1ab.pdb;
twwistPDB --exact new/4CEWx.pdb 5241 1188 3238 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/4CEW_tile_1a.pdb;
twwistPDB --exact new/4CEWx.pdb 5241 1188 3238 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/4CEW_tile_1b.pdb;
twwistPDB --exact new/4CEWx.pdb 5241 1188 3238 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/4CEW_tile_1c.pdb;
combinePDB --input1=new/super/4CEW_tile_1a.pdb --input2=new/super/4CEW_tile_1b.pdb --output=new/super/4CEW_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/4CEW_tile_1ab.pdb --input2=new/super/4CEW_tile_1c.pdb --output=new/super/4CEW_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/4CEW_tile_1a.pdb;
rm new/super/4CEW_tile_1b.pdb;
rm new/super/4CEW_tile_1c.pdb;
rm new/super/4CEW_tile_1ab.pdb;
twwistPDB --exact new/4Q4Yx.pdb 5372 1114 3222 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/4Q4Y_tile_1a.pdb;
twwistPDB --exact new/4Q4Yx.pdb 5372 1114 3222 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/4Q4Y_tile_1b.pdb;
twwistPDB --exact new/4Q4Yx.pdb 5372 1114 3222 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/4Q4Y_tile_1c.pdb;
combinePDB --input1=new/super/4Q4Y_tile_1a.pdb --input2=new/super/4Q4Y_tile_1b.pdb --output=new/super/4Q4Y_tile_1ab.pdb --chainid="ABCDEFGH";
combinePDB --input1=new/super/4Q4Y_tile_1ab.pdb --input2=new/super/4Q4Y_tile_1c.pdb --output=new/super/4Q4Y_tile_1.pdb --chainid="ABCDEFGHIJKL";
rm new/super/4Q4Y_tile_1a.pdb;
rm new/super/4Q4Y_tile_1b.pdb;
rm new/super/4Q4Y_tile_1c.pdb;
rm new/super/4Q4Y_tile_1ab.pdb;
twwistPDB --exact new/4WM7x.pdb 5123 1079 3154 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/4WM7_tile_1a.pdb;
twwistPDB --exact new/4WM7x.pdb 5123 1079 3154 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/4WM7_tile_1b.pdb;
twwistPDB --exact new/4WM7x.pdb 5123 1079 3154 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/4WM7_tile_1c.pdb;
combinePDB --input1=new/super/4WM7_tile_1a.pdb --input2=new/super/4WM7_tile_1b.pdb --output=new/super/4WM7_tile_1ab.pdb --chainid="ABCDEFGH" --map=new/super/4WM7_tile_1ab.map;
combinePDB --input1=new/super/4WM7_tile_1ab.pdb --input2=new/super/4WM7_tile_1c.pdb --output=new/super/4WM7_tile_1.pdb --chainid="ABCDEFGHIJKL" --map=new/super/4WM7_tile_1abc.map;
rm new/super/4WM7_tile_1b.pdb;
rm new/super/4WM7_tile_1c.pdb;
rm new/super/4WM7_tile_1ab.pdb;
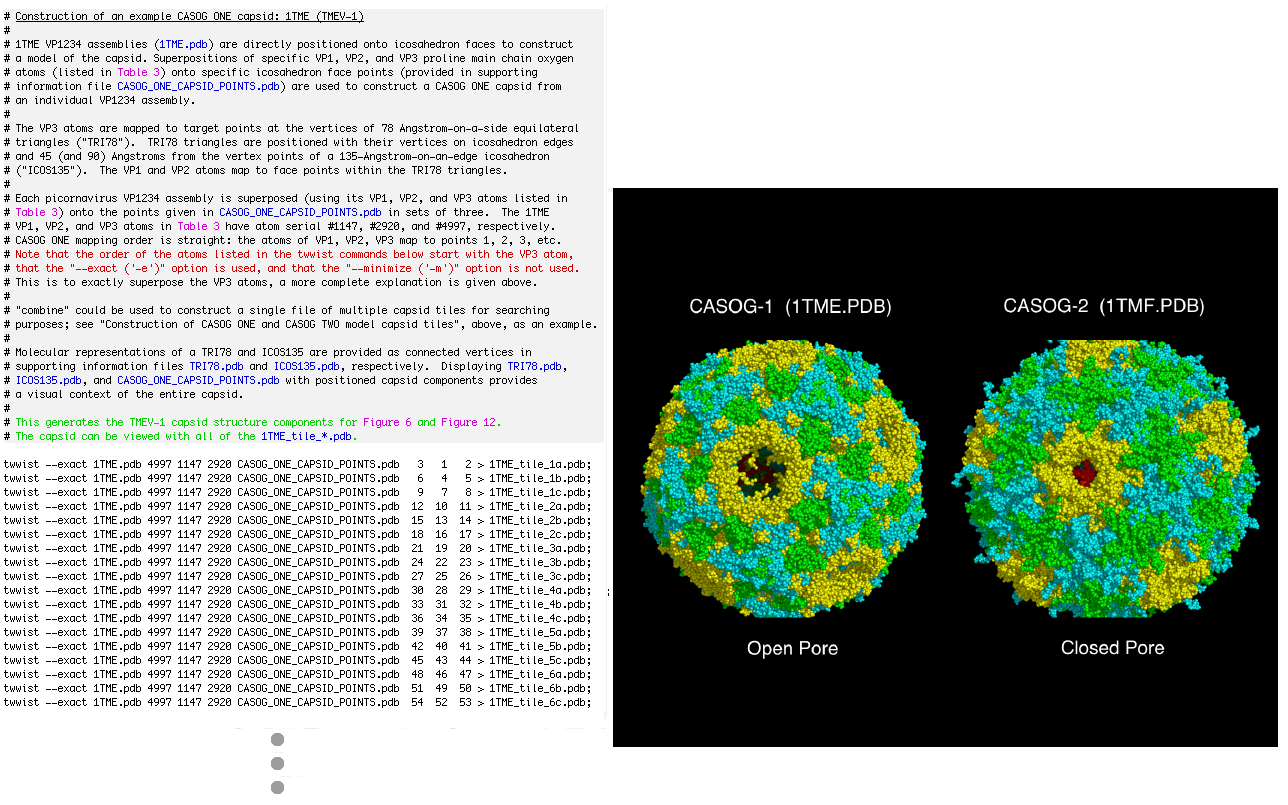
# Construction of an example CASOG ONE capsid: 1TME (TMEV-1)
#
# 1TME VP1234 assemblies (1TME.pdb) are directly positioned onto icosahedron faces to construct
# a model of the capsid. Superpositions of specific VP1, VP2, and VP3 proline main chain oxygen
# atoms (listed in Table 3) onto specific icosahedron face points (provided in supporting
# information file CASOG_ONE_CAPSID_POINTS.pdb) are used to construct a CASOG ONE capsid from
# an individual VP1234 assembly.
#
# The VP3 atoms are mapped to target points at the vertices of 78 Angstrom-on-a-side equilateral
# triangles ("TRI78"). TRI78 triangles are positioned with their vertices on icosahedron edges
# and 45 (and 90) Angstroms from the vertex points of a 135-Angstrom-on-an-edge icosahedron
# ("ICOS135"). The VP1 and VP2 atoms map to face points within the TRI78 triangles.
#
# Each picornavirus VP1234 assembly is superposed (using its VP1, VP2, and VP3 atoms listed in
# Table 3) onto the points given in CASOG_ONE_CAPSID_POINTS.pdb in sets of three. The 1TME
# VP1, VP2, and VP3 atoms in Table 3 have atom serial #1147, #2920, and #4997, respectively.
# CASOG ONE mapping order is straight: the atoms of VP1, VP2, VP3 map to points 1, 2, 3, etc.
# Note that the order of the atoms listed in the twwistPDB commands below start with the VP3 atom,
# that the "--exact ('-e')" option is used, and that the "--minimize ('-m')" option is not used.
# This is to exactly superpose the VP3 atoms, a more complete explanation is given above.
#
# "combinePDB" could be used to construct a single file of multiple capsid tiles for searching
# purposes; see "Construction of CASOG ONE and CASOG TWO model capsid tiles", above, as an example.
#
# Molecular representations of a TRI78 and ICOS135 are provided as connected vertices in
# supporting information files TRI78.pdb and ICOS135.pdb, respectively. Displaying TRI78.pdb,
# ICOS135.pdb, and CASOG_ONE_CAPSID_POINTS.pdb with positioned capsid components provides
# a visual context of the entire capsid.
#
# This generates the TMEV-1 capsid structure components for Figure 6 and Figure 12.
# The capsid can be viewed with all of the 1TME_tile_*.pdb.
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/1TME_capsid_tiles/1TME_tile_1a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/1TME_capsid_tiles/1TME_tile_1b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/1TME_capsid_tiles/1TME_tile_1c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 12 10 11 > new/super/1TME_capsid_tiles/1TME_tile_2a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 15 13 14 > new/super/1TME_capsid_tiles/1TME_tile_2b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 18 16 17 > new/super/1TME_capsid_tiles/1TME_tile_2c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 21 19 20 > new/super/1TME_capsid_tiles/1TME_tile_3a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 24 22 23 > new/super/1TME_capsid_tiles/1TME_tile_3b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 27 25 26 > new/super/1TME_capsid_tiles/1TME_tile_3c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 30 28 29 > new/super/1TME_capsid_tiles/1TME_tile_4a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 33 31 32 > new/super/1TME_capsid_tiles/1TME_tile_4b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 36 34 35 > new/super/1TME_capsid_tiles/1TME_tile_4c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 39 37 38 > new/super/1TME_capsid_tiles/1TME_tile_5a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 42 40 41 > new/super/1TME_capsid_tiles/1TME_tile_5b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 45 43 44 > new/super/1TME_capsid_tiles/1TME_tile_5c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 48 46 47 > new/super/1TME_capsid_tiles/1TME_tile_6a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 51 49 50 > new/super/1TME_capsid_tiles/1TME_tile_6b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 54 52 53 > new/super/1TME_capsid_tiles/1TME_tile_6c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 57 55 56 > new/super/1TME_capsid_tiles/1TME_tile_7a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 60 58 59 > new/super/1TME_capsid_tiles/1TME_tile_7b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 63 61 62 > new/super/1TME_capsid_tiles/1TME_tile_7c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 66 64 65 > new/super/1TME_capsid_tiles/1TME_tile_8a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 69 67 68 > new/super/1TME_capsid_tiles/1TME_tile_8b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 72 70 71 > new/super/1TME_capsid_tiles/1TME_tile_8c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 75 73 74 > new/super/1TME_capsid_tiles/1TME_tile_9a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 78 76 77 > new/super/1TME_capsid_tiles/1TME_tile_9b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 81 79 80 > new/super/1TME_capsid_tiles/1TME_tile_9c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 84 82 83 > new/super/1TME_capsid_tiles/1TME_tile_10a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 87 85 86 > new/super/1TME_capsid_tiles/1TME_tile_10b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 90 88 89 > new/super/1TME_capsid_tiles/1TME_tile_10c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 93 91 92 > new/super/1TME_capsid_tiles/1TME_tile_11a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 96 94 95 > new/super/1TME_capsid_tiles/1TME_tile_11b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 99 97 98 > new/super/1TME_capsid_tiles/1TME_tile_11c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 102 100 101 > new/super/1TME_capsid_tiles/1TME_tile_12a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 105 103 104 > new/super/1TME_capsid_tiles/1TME_tile_12b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 108 106 107 > new/super/1TME_capsid_tiles/1TME_tile_12c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 111 109 110 > new/super/1TME_capsid_tiles/1TME_tile_13a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 114 112 113 > new/super/1TME_capsid_tiles/1TME_tile_13b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 117 115 116 > new/super/1TME_capsid_tiles/1TME_tile_13c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 120 118 119 > new/super/1TME_capsid_tiles/1TME_tile_14a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 123 121 122 > new/super/1TME_capsid_tiles/1TME_tile_14b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 126 124 125 > new/super/1TME_capsid_tiles/1TME_tile_14c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 129 127 128 > new/super/1TME_capsid_tiles/1TME_tile_15a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 132 130 131 > new/super/1TME_capsid_tiles/1TME_tile_15b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 135 133 134 > new/super/1TME_capsid_tiles/1TME_tile_15c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 138 136 137 > new/super/1TME_capsid_tiles/1TME_tile_16a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 141 139 140 > new/super/1TME_capsid_tiles/1TME_tile_16b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 144 142 143 > new/super/1TME_capsid_tiles/1TME_tile_16c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 147 145 146 > new/super/1TME_capsid_tiles/1TME_tile_17a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 150 148 149 > new/super/1TME_capsid_tiles/1TME_tile_17b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 153 151 152 > new/super/1TME_capsid_tiles/1TME_tile_17c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 156 154 155 > new/super/1TME_capsid_tiles/1TME_tile_18a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 159 157 158 > new/super/1TME_capsid_tiles/1TME_tile_18b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 162 160 161 > new/super/1TME_capsid_tiles/1TME_tile_18c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 165 163 164 > new/super/1TME_capsid_tiles/1TME_tile_19a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 168 166 167 > new/super/1TME_capsid_tiles/1TME_tile_19b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 171 169 170 > new/super/1TME_capsid_tiles/1TME_tile_19c.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 174 172 173 > new/super/1TME_capsid_tiles/1TME_tile_20a.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 177 175 176 > new/super/1TME_capsid_tiles/1TME_tile_20b.pdb;
twwistPDB --exact new/1TMEx.pdb 4997 1147 2920 orig/CASOG_ONE_CAPSID_POINTS.pdb 180 178 179 > new/super/1TME_capsid_tiles/1TME_tile_20c.pdb;
# Construction of an example CASOG TWO capsid: 1TMF (TMEV-2)
#
# 1TMF VP1234 assemblies (1TMF.pdb) are directly positioned onto icosahedron faces to construct
# a model of the capsid. Superpositions of specific VP1, VP2, and VP3 proline main chain oxygen
# atoms (listed in Table 4) onto specific icosahedron face points (provided in supporting
# information file CASOG_TWO_CAPSID_POINTS.pdb) are used to construct a CASOG TWO capsid from
# an individual VP1234 assembly.
#
# The icosahedron face points that are used as superpositioning targets are points at the
# vertices of 26 Angstrom-on-a-side equilateral triangles ("TRI26"). TRI26 triangles are
# coplanar with, and positioned at the corners of, the TRI78 triangles (described above).
# (A molecular representation of TRI26 points are provided as connected vertices in
# CASOG_TWO_CAPSID_POINTS.pdb.)
#
# Each picornavirus VP1234 assembly is mapped (using its VP1, VP2, and VP3 atoms listed
# in Table 4) onto the points given in CASOG_TWO_CAPSID_POINTS.pdb in sets of three. The 1TMF
# VP1, VP2, and VP3 atoms in Table 4 have atom serial #1164, #3160, and #5234, respectively.
# CASOG TWO mapping order is straight: the atoms of VP1, VP2, VP3 map to points 1, 2, 3, etc.
#
# twwistPDB commands that map VP1234 assemblies to capsid face points in CASOG TWO orientation
# normally have the following format:
#
# twwistPDB initial_VP1234.pdb VP1 VP2 VP3 CASOG_TWO_CAPSID_POINTS.pdb N N+1 N+2 > VP1234_on_capsid.pdb
#
# The three commented twwistPDB commands, immediately below, are an example of normal twwistPDB commands.
#
# twwistPDB 1TMF.pdb 1164 3160 5234 orig/CASOG_TWO_CAPSID_POINTS.pdb 1 2 3 > 1TMF_tile_1a.pdb;
# twwistPDB 1TMF.pdb 1164 3160 5234 orig/CASOG_TWO_CAPSID_POINTS.pdb 4 5 6 > 1TMF_tile_1b.pdb;
# twwistPDB 1TMF.pdb 1164 3160 5234 orig/CASOG_TWO_CAPSID_POINTS.pdb 7 8 9 > 1TMF_tile_1c.pdb;
#
# These three twwistPDB commands place three VP1234 assemblies on the first capsid icosahedron face.
# "combinePDB" could be used to create a single capsid tiling piece from these reoriented VP1234
# assemblies. "combinePDB" could be further used to construct a single file of multiple capsid tiles
# for searching purposes. See "Construction of CASOG ONE and CASOG TWO model capsid tiles", above,
# as an example.
#
# The actual commands used, just below, are special in that they prioritize VP3 atom placement
# instead of minimizing the sum of all atom placement error. The order of the atoms listed in
# the twwistPDB commands below starts with the VP3 atom, the "--exact ('-e')" option is used,
# and the "--minimize ('-m')" option is not used. (This combination of options specifies that
# the first atoms will be exactly superposed, and that any existing differences in distances
# between the pairs of specified atoms in the two different molecules will not be minimized.
# The VP3 atom is the pivot point between CASOG ONE and CASOG TWO orientations.)
# The following format is used:
#
# twwistPDB --exact initVP1234.pdb VP3 VP1 VP2 CASOG_TWO_CAPSID_POINTS.pdb N+2 N N+1 > capsidVP1234.pdb
#
# This generates the TMEV-2 capsid structure components for Figure 6 and Figure 12.
# The capsid can be viewed with all of the 1TMF_tile_*.pdb.
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 3 1 2 > new/super/1TMF_capsid_tiles/1TMF_tile_1a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 6 4 5 > new/super/1TMF_capsid_tiles/1TMF_tile_1b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 9 7 8 > new/super/1TMF_capsid_tiles/1TMF_tile_1c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 12 10 11 > new/super/1TMF_capsid_tiles/1TMF_tile_2a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 15 13 14 > new/super/1TMF_capsid_tiles/1TMF_tile_2b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 18 16 17 > new/super/1TMF_capsid_tiles/1TMF_tile_2c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 21 19 20 > new/super/1TMF_capsid_tiles/1TMF_tile_3a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 24 22 23 > new/super/1TMF_capsid_tiles/1TMF_tile_3b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 27 25 26 > new/super/1TMF_capsid_tiles/1TMF_tile_3c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 30 28 29 > new/super/1TMF_capsid_tiles/1TMF_tile_4a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 33 31 32 > new/super/1TMF_capsid_tiles/1TMF_tile_4b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 36 34 35 > new/super/1TMF_capsid_tiles/1TMF_tile_4c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 39 37 38 > new/super/1TMF_capsid_tiles/1TMF_tile_5a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 42 40 41 > new/super/1TMF_capsid_tiles/1TMF_tile_5b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 45 43 44 > new/super/1TMF_capsid_tiles/1TMF_tile_5c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 48 46 47 > new/super/1TMF_capsid_tiles/1TMF_tile_6a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 51 49 50 > new/super/1TMF_capsid_tiles/1TMF_tile_6b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 54 52 53 > new/super/1TMF_capsid_tiles/1TMF_tile_6c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 57 55 56 > new/super/1TMF_capsid_tiles/1TMF_tile_7a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 60 58 59 > new/super/1TMF_capsid_tiles/1TMF_tile_7b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 63 61 62 > new/super/1TMF_capsid_tiles/1TMF_tile_7c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 66 64 65 > new/super/1TMF_capsid_tiles/1TMF_tile_8a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 69 67 68 > new/super/1TMF_capsid_tiles/1TMF_tile_8b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 72 70 71 > new/super/1TMF_capsid_tiles/1TMF_tile_8c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 75 73 74 > new/super/1TMF_capsid_tiles/1TMF_tile_9a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 78 76 77 > new/super/1TMF_capsid_tiles/1TMF_tile_9b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 81 79 80 > new/super/1TMF_capsid_tiles/1TMF_tile_9c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 84 82 83 > new/super/1TMF_capsid_tiles/1TMF_tile_10a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 87 85 86 > new/super/1TMF_capsid_tiles/1TMF_tile_10b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 90 88 89 > new/super/1TMF_capsid_tiles/1TMF_tile_10c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 93 91 92 > new/super/1TMF_capsid_tiles/1TMF_tile_11a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 96 94 95 > new/super/1TMF_capsid_tiles/1TMF_tile_11b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 99 97 98 > new/super/1TMF_capsid_tiles/1TMF_tile_11c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 102 100 101 > new/super/1TMF_capsid_tiles/1TMF_tile_12a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 105 103 104 > new/super/1TMF_capsid_tiles/1TMF_tile_12b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 108 106 107 > new/super/1TMF_capsid_tiles/1TMF_tile_12c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 111 109 110 > new/super/1TMF_capsid_tiles/1TMF_tile_13a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 114 112 113 > new/super/1TMF_capsid_tiles/1TMF_tile_13b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 117 115 116 > new/super/1TMF_capsid_tiles/1TMF_tile_13c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 120 118 119 > new/super/1TMF_capsid_tiles/1TMF_tile_14a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 123 121 122 > new/super/1TMF_capsid_tiles/1TMF_tile_14b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 126 124 125 > new/super/1TMF_capsid_tiles/1TMF_tile_14c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 129 127 128 > new/super/1TMF_capsid_tiles/1TMF_tile_15a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 132 130 131 > new/super/1TMF_capsid_tiles/1TMF_tile_15b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 135 133 134 > new/super/1TMF_capsid_tiles/1TMF_tile_15c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 138 136 137 > new/super/1TMF_capsid_tiles/1TMF_tile_16a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 141 139 140 > new/super/1TMF_capsid_tiles/1TMF_tile_16b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 144 142 143 > new/super/1TMF_capsid_tiles/1TMF_tile_16c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 147 145 146 > new/super/1TMF_capsid_tiles/1TMF_tile_17a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 150 148 149 > new/super/1TMF_capsid_tiles/1TMF_tile_17b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 153 151 152 > new/super/1TMF_capsid_tiles/1TMF_tile_17c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 156 154 155 > new/super/1TMF_capsid_tiles/1TMF_tile_18a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 159 157 158 > new/super/1TMF_capsid_tiles/1TMF_tile_18b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 162 160 161 > new/super/1TMF_capsid_tiles/1TMF_tile_18c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 165 163 164 > new/super/1TMF_capsid_tiles/1TMF_tile_19a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 168 166 167 > new/super/1TMF_capsid_tiles/1TMF_tile_19b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 171 169 170 > new/super/1TMF_capsid_tiles/1TMF_tile_19c.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 174 172 173 > new/super/1TMF_capsid_tiles/1TMF_tile_20a.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 177 175 176 > new/super/1TMF_capsid_tiles/1TMF_tile_20b.pdb;
twwistPDB --exact new/1TMFx.pdb 5234 1164 3160 orig/CASOG_TWO_CAPSID_POINTS.pdb 180 178 179 > new/super/1TMF_capsid_tiles/1TMF_tile_20c.pdb;
# Construction of human myelin P2 protein (2WUT) trimers and CRABP (1CBS) trimers
#
# Human myelin P2 protein (2WUT.pdb) and CRABP monomers (1CBS.pdb) were separately superposed
# onto combined CASOG ONE and CASOG TWO tiling points (provided in supporting information file
# PICORNAVIRUS_TILING_POINTS.pdb) using atoms listed in Table 5.
#
# This generates two structures, 2WUT_TRIMER_table5.pdb and 1CBS_TRIMER_table5.pdb, for
# Figure 7 Panels C and D, Figure 8 Columns 0 and 1, Figure 9 Panels A and B, and Figure 15 Row A.
twwistPDB --exact --output=new/super/2WUT_on_TRIMER_1.pdb new/2WUTx.pdb 877 330 506 orig/PICORNAVIRUS_TILING_POINTS.pdb 21 22 23;
twwistPDB --exact --output=new/super/2WUT_on_TRIMER_2.pdb new/2WUTx.pdb 877 330 506 orig/PICORNAVIRUS_TILING_POINTS.pdb 24 25 26;
twwistPDB --exact --output=new/super/2WUT_on_TRIMER_3.pdb new/2WUTx.pdb 877 330 506 orig/PICORNAVIRUS_TILING_POINTS.pdb 27 28 29;
combinePDB --input1=new/super/2WUT_on_TRIMER_1.pdb --input2=new/super/2WUT_on_TRIMER_2.pdb --output=new/super/2WUT_on_TRIMER_12.pdb;
combinePDB --input1=new/super/2WUT_on_TRIMER_12.pdb --input2=new/super/2WUT_on_TRIMER_3.pdb --output=new/super/2WUT_TRIMER_table5.pdb;
rm new/super/2WUT_on_TRIMER_1.pdb;
rm new/super/2WUT_on_TRIMER_2.pdb;
rm new/super/2WUT_on_TRIMER_3.pdb;
rm new/super/2WUT_on_TRIMER_12.pdb;
twwistPDB --exact --output=new/super/1CBS_on_TRIMER_1.pdb new/1CBSx.pdb 871 324 495 orig/PICORNAVIRUS_TILING_POINTS.pdb 21 22 23;
twwistPDB --exact --output=new/super/1CBS_on_TRIMER_2.pdb new/1CBSx.pdb 871 324 495 orig/PICORNAVIRUS_TILING_POINTS.pdb 24 25 26;
twwistPDB --exact --output=new/super/1CBS_on_TRIMER_3.pdb new/1CBSx.pdb 871 324 495 orig/PICORNAVIRUS_TILING_POINTS.pdb 27 28 29;
combinePDB --input1=new/super/1CBS_on_TRIMER_1.pdb --input2=new/super/1CBS_on_TRIMER_2.pdb --output=new/super/1CBS_on_TRIMER_12.pdb;
combinePDB --input1=new/super/1CBS_on_TRIMER_12.pdb --input2=new/super/1CBS_on_TRIMER_3.pdb --output=new/super/1CBS_TRIMER_table5.pdb;
rm new/super/1CBS_on_TRIMER_1.pdb;
rm new/super/1CBS_on_TRIMER_2.pdb;
rm new/super/1CBS_on_TRIMER_3.pdb;
rm new/super/1CBS_on_TRIMER_12.pdb;
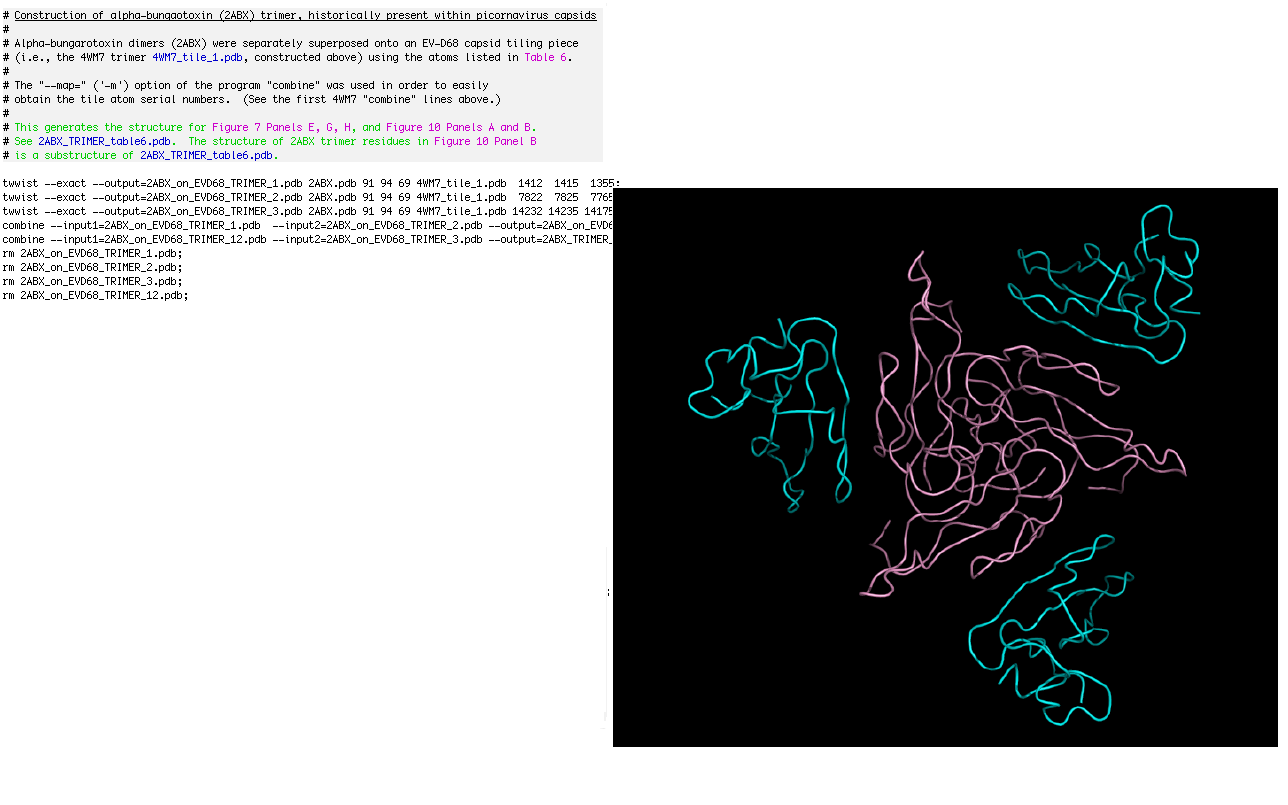
# Construction of alpha-bungaotoxin (2ABX) trimer, historically present within picornavirus capsids
#
# Alpha-bungarotoxin dimers (2ABX.pdb) were separately superposed onto an EV-D68 capsid tiling piece
# (i.e., the 4WM7 trimer 4WM7_tile_1.pdb, constructed above) using the atoms listed in Table 6.
#
# The "--map=" ('-m') option of the program "combinePDB" was used in order to easily
# obtain the tile atom serial numbers. (See the first 4WM7 "combinePDB" lines above.)
#
# This generates the structure for Figure 7 Panels E, G, H, and Figure 10 Panels A and B.
# See 2ABX_TRIMER_table6.pdb. The structure of 2ABX trimer residues in Figure 10 Panel B
# is a substructure of 2ABX_TRIMER_table6.pdb.
twwistPDB --exact --output=new/super/2ABX_on_EVD68_TRIMER_1.pdb new/2ABXx.pdb 91 94 69 new/super/4WM7_tile_1.pdb 1412 1415 1355;
twwistPDB --exact --output=new/super/2ABX_on_EVD68_TRIMER_2.pdb new/2ABXx.pdb 91 94 69 new/super/4WM7_tile_1.pdb 7672 7675 7615;
twwistPDB --exact --output=new/super/2ABX_on_EVD68_TRIMER_3.pdb new/2ABXx.pdb 91 94 69 new/super/4WM7_tile_1.pdb 13932 13935 13875;
combinePDB --input1=new/super/2ABX_on_EVD68_TRIMER_1.pdb --input2=new/super/2ABX_on_EVD68_TRIMER_2.pdb --output=new/super/2ABX_on_EVD68_TRIMER_12.pdb;
combinePDB --input1=new/super/2ABX_on_EVD68_TRIMER_12.pdb --input2=new/super/2ABX_on_EVD68_TRIMER_3.pdb --output=new/super/2ABX_TRIMER_table6.pdb;
rm new/super/2ABX_on_EVD68_TRIMER_1.pdb;
rm new/super/2ABX_on_EVD68_TRIMER_2.pdb;
rm new/super/2ABX_on_EVD68_TRIMER_3.pdb;
rm new/super/2ABX_on_EVD68_TRIMER_12.pdb;
# New PDB files are made for monograph figures.
#
# PDB files should now have been made for each superposition in the monograph.
#
# To provide focus for specific features, monograph structural figures show a subset,
# a combination, or a subset of a combination of the PDB files showing molecular superpositions.
# New subdirectories are made for new files.
#
# PDB files for individual figure panels will now be made and written to 'figs',
# a subdirectory of 'new'. These PDB files will be named by figure, panel, and content.
# Where multiple PDB files are created for a single panel (e.g., to help display different
# colors or styles), the PDB filenames will also include a PDB count for that panel (e.g., "1of4").
# Where multiple panels share the same PDB files (e.g., panel A shows an outer view and panel B
# shows an inner view), a single set of PDB files will created that have filenames that include
# multiple panel specifications (e.g., "panels_A_B").
mkdir new/figs;
mkdir new/figs/fig4;
mkdir new/figs/fig5;
mkdir new/figs/fig6;
mkdir new/figs/fig7;
mkdir new/figs/fig8;
mkdir new/figs/fig9;
mkdir new/figs/fig10;
mkdir new/figs/fig11;
mkdir new/figs/fig12;
mkdir new/figs/fig13;
mkdir new/figs/fig14;
mkdir new/figs/fig15;
# Figure 4. Superposition of VP1-VP2-VP3 picornavirus protein assemblies
# showing CASOG-1 and CASOG-2 orientations.
# Copy and name PDBs for Figure 4 Panels B1 - Q1
# (Copy aligned individual VP1234 structures.)
cp new/super/1AYM_on_VP1.pdb new/figs/fig4/figure_4_panel_B1-1AYM_VP1234.pdb;
cp new/super/1BBT_on_VP1.pdb new/figs/fig4/figure_4_panel_C1-1BBT_VP1234.pdb;
cp new/super/1BEV_on_VP1.pdb new/figs/fig4/figure_4_panel_D1-1BEV_VP1234.pdb;
cp new/super/1D4M_on_VP1.pdb new/figs/fig4/figure_4_panel_E1-1D4M_VP1234.pdb;
cp new/super/1EAH_on_VP1.pdb new/figs/fig4/figure_4_panel_F1-1EAH_VP1234.pdb;
cp new/super/1HXS_on_VP1.pdb new/figs/fig4/figure_4_panel_G1-1HXS_VP1234.pdb;
cp new/super/1OOP_on_VP1.pdb new/figs/fig4/figure_4_panel_H1-1OOP_VP1234.pdb;
cp new/super/1PIV_on_VP1.pdb new/figs/fig4/figure_4_panel_I1-1PIV_VP1234.pdb;
cp new/super/1TME_on_VP1.pdb new/figs/fig4/figure_4_panel_J1-1TME_VP1234.pdb;
cp new/super/1TMF_on_VP1.pdb new/figs/fig4/figure_4_panel_K1-1TMF_VP1234.pdb;
cp new/super/1V9U_on_VP1.pdb new/figs/fig4/figure_4_panel_L1-1V9U_VP1234.pdb;
cp new/super/1Z7S_on_VP1.pdb new/figs/fig4/figure_4_panel_M1-1Z7S_VP1234.pdb;
cp new/super/3VBH_on_VP1.pdb new/figs/fig4/figure_4_panel_N1-3VBH_VP1234.pdb;
cp new/super/4CEW_on_VP1.pdb new/figs/fig4/figure_4_panel_O1-4CEW_VP1234.pdb;
cp new/super/4Q4Y_on_VP1.pdb new/figs/fig4/figure_4_panel_P1-4Q4Y_VP1234.pdb;
cp new/super/4WM7_on_VP1.pdb new/figs/fig4/figure_4_panel_Q1-4WM7_VP1234.pdb;
# Make PDBs for Figure 4 Panel A1.
# (Combine aligned individual VP1234 structures.)
#
# Note that two PDBs ("_1of2" and "_2of2") are necessary to hold structures for Figure 4 Panel A1
# as the limit of the number of atoms per PDB (99999) gets exceeded if only one PDB is used.
combinePDB --input1=new/super/1AYM_on_VP1.pdb \
--input2=new/super/1BBT_on_VP1.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/1BEV_on_VP1.pdb \
--input2=new/super/1D4M_on_VP1.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/1EAH_on_VP1.pdb \
--input2=new/super/1HXS_on_VP1.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/1OOP_on_VP1.pdb \
--input2=new/super/1PIV_on_VP1.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A1_1of2-COMBINED_VP1234.pdb;
combinePDB --input1=new/super/1TME_on_VP1.pdb \
--input2=new/super/1TMF_on_VP1.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/1V9U_on_VP1.pdb \
--input2=new/super/1Z7S_on_VP1.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/3VBH_on_VP1.pdb \
--input2=new/super/4CEW_on_VP1.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/4Q4Y_on_VP1.pdb \
--input2=new/super/4WM7_on_VP1.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A1_2of2-COMBINED_VP1234.pdb;
# Copy and name PDBs for Figure 4 Panels B2 - Q2
# (Copy aligned individual VP1234 trimers,"3xVP1234 capsid tiles".)
cp new/super/1AYM_tile_1.pdb new/figs/fig4/figure_4_panel_B2-1AYM_VP1234_TRIMER.pdb;
cp new/super/1BBT_tile_1.pdb new/figs/fig4/figure_4_panel_C2-1BBT_VP1234_TRIMER.pdb;
cp new/super/1BEV_tile_1.pdb new/figs/fig4/figure_4_panel_D2-1BEV_VP1234_TRIMER.pdb;
cp new/super/1D4M_tile_1.pdb new/figs/fig4/figure_4_panel_E2-1D4M_VP1234_TRIMER.pdb;
cp new/super/1EAH_tile_1.pdb new/figs/fig4/figure_4_panel_F2-1EAH_VP1234_TRIMER.pdb;
cp new/super/1HXS_tile_1.pdb new/figs/fig4/figure_4_panel_G2-1HXS_VP1234_TRIMER.pdb;
cp new/super/1OOP_tile_1.pdb new/figs/fig4/figure_4_panel_H2-1OOP_VP1234_TRIMER.pdb;
cp new/super/1PIV_tile_1.pdb new/figs/fig4/figure_4_panel_I2-1PIV_VP1234_TRIMER.pdb;
cp new/super/1TME_tile_1.pdb new/figs/fig4/figure_4_panel_J2-1TME_VP1234_TRIMER.pdb;
cp new/super/1TMF_tile_1.pdb new/figs/fig4/figure_4_panel_K2-1TMF_VP1234_TRIMER.pdb;
cp new/super/1V9U_tile_1.pdb new/figs/fig4/figure_4_panel_L2-1V9U_VP1234_TRIMER.pdb;
cp new/super/1Z7S_tile_1.pdb new/figs/fig4/figure_4_panel_M2-1Z7S_VP1234_TRIMER.pdb;
cp new/super/3VBH_tile_1.pdb new/figs/fig4/figure_4_panel_N2-3VBH_VP1234_TRIMER.pdb;
cp new/super/4CEW_tile_1.pdb new/figs/fig4/figure_4_panel_O2-4CEW_VP1234_TRIMER.pdb;
cp new/super/4Q4Y_tile_1.pdb new/figs/fig4/figure_4_panel_P2-4Q4Y_VP1234_TRIMER.pdb;
cp new/super/4WM7_tile_1.pdb new/figs/fig4/figure_4_panel_Q2-4WM7_VP1234_TRIMER.pdb;
# Make PDBs for Figure 4 Panel A2.
# (Combine aligned individual VP1234 trimers,"3xVP1234 capsid tiles".)
#
# Note that four PDBs("_1of4","_2of4","_3of4","_4of4") are necessary to hold structures for Figure 4
# Panel A2 as the limit of the number of atoms per PDB (99999) gets exceeded if only one PDB is used.
combinePDB --input1=new/super/1AYM_tile_1.pdb \
--input2=new/super/1BBT_tile_1.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/1BEV_tile_1.pdb \
--input2=new/super/1D4M_tile_1.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/super/1EAH_tile_1.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A2_1of4-COMBINED_VP1234_TRIMER.pdb;
combinePDB --input1=new/super/1HXS_tile_1.pdb \
--input2=new/super/1OOP_tile_1.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/1PIV_tile_1.pdb \
--input2=new/super/1TME_tile_1.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/super/1TMF_tile_1.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A2_2of4-COMBINED_VP1234_TRIMER.pdb;
combinePDB --input1=new/super/1V9U_tile_1.pdb \
--input2=new/super/1Z7S_tile_1.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/super/3VBH_tile_1.pdb \
--input2=new/super/4CEW_tile_1.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/super/4Q4Y_tile_1.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A2_3of4-COMBINED_VP1234_TRIMER.pdb;
cp new/super/4WM7_tile_1.pdb new/figs/fig4/figure_4_panel_A2_4of4-COMBINED_VP1234_TRIMER.pdb;
# Make PDBs for Figure 4 Panels B3 - Q3
# (Remove chains '2', '3', and '4' from aligned individual VP1234 trimers.)
#
# When the 'XXXX_tile_1' PDB files were created using combinePDB
# (see above "Construction of CASOG ONE and CASOG TWO model capsid tiles" above),
# the option --chainid="ABCDEFGH" was used to:
#
# (i) give each VP1234 within the 3xVP1234 tile a separate chain identifier, and
# (ii) standardize the chain identifiers
# (some VP1234 PDBs were originally '1','2','3','4', others 'A','B','C','D').
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# The following removes chains '2' ('B', 'F', 'J'), '3' ('C', 'G', 'K'), and
# '4' ('D', 'H', 'L') from aligned individual VP1234 trimers.
stripPDB --input=new/super/1AYM_tile_1.pdb --output=new/figs/fig4/figure_4_panel_B3-1AYM_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1BBT_tile_1.pdb --output=new/figs/fig4/figure_4_panel_C3-1BBT_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1BEV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_D3-1BEV_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1D4M_tile_1.pdb --output=new/figs/fig4/figure_4_panel_E3-1D4M_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1EAH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_F3-1EAH_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1HXS_tile_1.pdb --output=new/figs/fig4/figure_4_panel_G3-1HXS_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1OOP_tile_1.pdb --output=new/figs/fig4/figure_4_panel_H3-1OOP_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1PIV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_I3-1PIV_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1TME_tile_1.pdb --output=new/figs/fig4/figure_4_panel_J3-1TME_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1TMF_tile_1.pdb --output=new/figs/fig4/figure_4_panel_K3-1TMF_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1V9U_tile_1.pdb --output=new/figs/fig4/figure_4_panel_L3-1V9U_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/1Z7S_tile_1.pdb --output=new/figs/fig4/figure_4_panel_M3-1Z7S_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/3VBH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_N3-3VBH_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/4CEW_tile_1.pdb --output=new/figs/fig4/figure_4_panel_O3-4CEW_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/4Q4Y_tile_1.pdb --output=new/figs/fig4/figure_4_panel_P3-4Q4Y_VP1_TRIMER.pdb --remove="BFJCGKDHL";
stripPDB --input=new/super/4WM7_tile_1.pdb --output=new/figs/fig4/figure_4_panel_Q3-4WM7_VP1_TRIMER.pdb --remove="BFJCGKDHL";
# Make PDBs for Figure 4 Panel A3.
# (Combine aligned individual VP1 trimers.)
#
# Note that two PDBs ("_1of2" and "_2of2") are necessary to hold structures for Figure 4 Panel A3
# as the limit of the number of atoms per PDB (99999) gets exceeded if only one PDB is used.
combinePDB --input1=new/figs/fig4/figure_4_panel_B3-1AYM_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_C3-1BBT_VP1_TRIMER.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_D3-1BEV_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_E3-1D4M_VP1_TRIMER.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_F3-1EAH_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_G3-1HXS_VP1_TRIMER.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_H3-1OOP_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_I3-1PIV_VP1_TRIMER.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A3_1of2-COMBINED_VP1_TRIMER.pdb;
combinePDB --input1=new/figs/fig4/figure_4_panel_J3-1TME_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_K3-1TMF_VP1_TRIMER.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_L3-1V9U_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_M3-1Z7S_VP1_TRIMER.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_N3-3VBH_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_O3-4CEW_VP1_TRIMER.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_P3-4Q4Y_VP1_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_Q3-4WM7_VP1_TRIMER.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A3_2of2-COMBINED_VP1_TRIMER.pdb;
# Make PDBs for Figure 4 Panels B4 - Q4.
# (Remove chains '1', '3', and '4' from aligned individual VP1234 trimers.)
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# The following removes chains '1' ('A', 'E', 'I'), '3' ('C', 'G', 'K'), and
# '4' ('D', 'H', 'L') from aligned individual VP1234 trimers.
stripPDB --input=new/super/1AYM_tile_1.pdb --output=new/figs/fig4/figure_4_panel_B4-1AYM_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1BBT_tile_1.pdb --output=new/figs/fig4/figure_4_panel_C4-1BBT_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1BEV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_D4-1BEV_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1D4M_tile_1.pdb --output=new/figs/fig4/figure_4_panel_E4-1D4M_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1EAH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_F4-1EAH_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1HXS_tile_1.pdb --output=new/figs/fig4/figure_4_panel_G4-1HXS_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1OOP_tile_1.pdb --output=new/figs/fig4/figure_4_panel_H4-1OOP_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1PIV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_I4-1PIV_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1TME_tile_1.pdb --output=new/figs/fig4/figure_4_panel_J4-1TME_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1TMF_tile_1.pdb --output=new/figs/fig4/figure_4_panel_K4-1TMF_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1V9U_tile_1.pdb --output=new/figs/fig4/figure_4_panel_L4-1V9U_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/1Z7S_tile_1.pdb --output=new/figs/fig4/figure_4_panel_M4-1Z7S_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/3VBH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_N4-3VBH_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/4CEW_tile_1.pdb --output=new/figs/fig4/figure_4_panel_O4-4CEW_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/4Q4Y_tile_1.pdb --output=new/figs/fig4/figure_4_panel_P4-4Q4Y_VP2_TRIMER.pdb --remove="AEICGKDHL";
stripPDB --input=new/super/4WM7_tile_1.pdb --output=new/figs/fig4/figure_4_panel_Q4-4WM7_VP2_TRIMER.pdb --remove="AEICGKDHL";
# Make PDB for Figure 4 Panel A4
# (Combine aligned individual VP2 trimers.)
combinePDB --input1=new/figs/fig4/figure_4_panel_B4-1AYM_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_C4-1BBT_VP2_TRIMER.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_D4-1BEV_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_E4-1D4M_VP2_TRIMER.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_F4-1EAH_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_G4-1HXS_VP2_TRIMER.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_H4-1OOP_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_I4-1PIV_VP2_TRIMER.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_J4-1TME_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_K4-1TMF_VP2_TRIMER.pdb \
--output=new/tmp5.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_L4-1V9U_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_M4-1Z7S_VP2_TRIMER.pdb \
--output=new/tmp6.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_N4-3VBH_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_O4-4CEW_VP2_TRIMER.pdb \
--output=new/tmp7.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_P4-4Q4Y_VP2_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_Q4-4WM7_VP2_TRIMER.pdb \
--output=new/tmp8.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--output=new/tmp1234.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp5.pdb \
--input2=new/tmp6.pdb \
--output=new/tmp56.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp7.pdb \
--input2=new/tmp8.pdb \
--output=new/tmp78.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp56.pdb \
--input2=new/tmp78.pdb \
--output=new/tmp5678.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1234.pdb \
--input2=new/tmp5678.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A4-COMBINED_VP2_TRIMER.pdb;
# Make PDBs for Figure 4 Panels B5 - Q5.
# (Remove chains '1', '2', and '4' from aligned individual VP1234 trimers.)
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# The following removes chains '1' ('A', 'E', 'I'), '2' ('B', 'F', 'J'), and
# '4' ('D', 'H', 'L') from aligned individual VP1234 trimers.
stripPDB --input=new/super/1AYM_tile_1.pdb --output=new/figs/fig4/figure_4_panel_B5-1AYM_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1BBT_tile_1.pdb --output=new/figs/fig4/figure_4_panel_C5-1BBT_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1BEV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_D5-1BEV_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1D4M_tile_1.pdb --output=new/figs/fig4/figure_4_panel_E5-1D4M_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1EAH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_F5-1EAH_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1HXS_tile_1.pdb --output=new/figs/fig4/figure_4_panel_G5-1HXS_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1OOP_tile_1.pdb --output=new/figs/fig4/figure_4_panel_H5-1OOP_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1PIV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_I5-1PIV_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1TME_tile_1.pdb --output=new/figs/fig4/figure_4_panel_J5-1TME_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1TMF_tile_1.pdb --output=new/figs/fig4/figure_4_panel_K5-1TMF_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1V9U_tile_1.pdb --output=new/figs/fig4/figure_4_panel_L5-1V9U_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/1Z7S_tile_1.pdb --output=new/figs/fig4/figure_4_panel_M5-1Z7S_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/3VBH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_N5-3VBH_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/4CEW_tile_1.pdb --output=new/figs/fig4/figure_4_panel_O5-4CEW_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/4Q4Y_tile_1.pdb --output=new/figs/fig4/figure_4_panel_P5-4Q4Y_VP3_TRIMER.pdb --remove="AEIBFJDHL";
stripPDB --input=new/super/4WM7_tile_1.pdb --output=new/figs/fig4/figure_4_panel_Q5-4WM7_VP3_TRIMER.pdb --remove="AEIBFJDHL";
# Make PDBs for Figure 4 Panel A5.
# (Combine aligned individual VP3 trimers.)
combinePDB --input1=new/figs/fig4/figure_4_panel_B5-1AYM_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_C5-1BBT_VP3_TRIMER.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_D5-1BEV_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_E5-1D4M_VP3_TRIMER.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_F5-1EAH_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_G5-1HXS_VP3_TRIMER.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_H5-1OOP_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_I5-1PIV_VP3_TRIMER.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_J5-1TME_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_K5-1TMF_VP3_TRIMER.pdb \
--output=new/tmp5.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_L5-1V9U_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_M5-1Z7S_VP3_TRIMER.pdb \
--output=new/tmp6.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_N5-3VBH_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_O5-4CEW_VP3_TRIMER.pdb \
--output=new/tmp7.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_P5-4Q4Y_VP3_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_Q5-4WM7_VP3_TRIMER.pdb \
--output=new/tmp8.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--output=new/tmp1234.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp5.pdb \
--input2=new/tmp6.pdb \
--output=new/tmp56.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp7.pdb \
--input2=new/tmp8.pdb \
--output=new/tmp78.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp56.pdb \
--input2=new/tmp78.pdb \
--output=new/tmp5678.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1234.pdb \
--input2=new/tmp5678.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A5-COMBINED_VP3_TRIMER.pdb;
# Make PDBs for Figure 4 Panels B6 - Q6.
# (Remove chains '1', '2', and '3' from aligned individual VP1234 trimers.)
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# The following removes chains '1' ('A', 'E', 'I'), '2' ('B', 'F', 'J'), and
# '3' ('C', 'G', 'K') from aligned individual VP1234 trimers.
stripPDB --input=new/super/1AYM_tile_1.pdb --output=new/figs/fig4/figure_4_panel_B6-1AYM_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1BBT_tile_1.pdb --output=new/figs/fig4/figure_4_panel_C6-1BBT_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1BEV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_D6-1BEV_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1D4M_tile_1.pdb --output=new/figs/fig4/figure_4_panel_E6-1D4M_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1EAH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_F6-1EAH_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1HXS_tile_1.pdb --output=new/figs/fig4/figure_4_panel_G6-1HXS_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1OOP_tile_1.pdb --output=new/figs/fig4/figure_4_panel_H6-1OOP_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1PIV_tile_1.pdb --output=new/figs/fig4/figure_4_panel_I6-1PIV_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1TME_tile_1.pdb --output=new/figs/fig4/figure_4_panel_J6-1TME_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1TMF_tile_1.pdb --output=new/figs/fig4/figure_4_panel_K6-1TMF_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1V9U_tile_1.pdb --output=new/figs/fig4/figure_4_panel_L6-1V9U_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/1Z7S_tile_1.pdb --output=new/figs/fig4/figure_4_panel_M6-1Z7S_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/3VBH_tile_1.pdb --output=new/figs/fig4/figure_4_panel_N6-3VBH_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/4CEW_tile_1.pdb --output=new/figs/fig4/figure_4_panel_O6-4CEW_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/4Q4Y_tile_1.pdb --output=new/figs/fig4/figure_4_panel_P6-4Q4Y_VP4_TRIMER.pdb --remove="AEIBFJCGK";
stripPDB --input=new/super/4WM7_tile_1.pdb --output=new/figs/fig4/figure_4_panel_Q6-4WM7_VP4_TRIMER.pdb --remove="AEIBFJCGK";
# Make PDBs for Figure 4 Panel A6.
# (Combine aligned individual VP4 trimers.)
combinePDB --input1=new/figs/fig4/figure_4_panel_B6-1AYM_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_C6-1BBT_VP4_TRIMER.pdb \
--output=new/tmp1.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_D6-1BEV_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_E6-1D4M_VP4_TRIMER.pdb \
--output=new/tmp2.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_F6-1EAH_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_G6-1HXS_VP4_TRIMER.pdb \
--output=new/tmp3.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_H6-1OOP_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_I6-1PIV_VP4_TRIMER.pdb \
--output=new/tmp4.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_J6-1TME_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_K6-1TMF_VP4_TRIMER.pdb \
--output=new/tmp5.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_L6-1V9U_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_M6-1Z7S_VP4_TRIMER.pdb \
--output=new/tmp6.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_N6-3VBH_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_O6-4CEW_VP4_TRIMER.pdb \
--output=new/tmp7.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/figs/fig4/figure_4_panel_P6-4Q4Y_VP4_TRIMER.pdb \
--input2=new/figs/fig4/figure_4_panel_Q6-4WM7_VP4_TRIMER.pdb \
--output=new/tmp8.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--output=new/tmp1234.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp5.pdb \
--input2=new/tmp6.pdb \
--output=new/tmp56.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp7.pdb \
--input2=new/tmp8.pdb \
--output=new/tmp78.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp56.pdb \
--input2=new/tmp78.pdb \
--output=new/tmp5678.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1234.pdb \
--input2=new/tmp5678.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig4/figure_4_panel_A6-COMBINED_VP4_TRIMER.pdb;
# Figure 5. Model picornavirus capsid construction for the CASOG-1 and CASOG-2 proteins.
# (Combine PDBs of geometric points.)
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panels_A_D_E_F_J_K_L_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panels_A_D_E_F_J_K_L_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
stripPDB --input=orig/PICORNAVIRUS_TILING_POINTS.pdb --remove="3" \
--output=new/figs/fig5/figure_5_panels_A_D_E_F_J_K_L_3of3-CASOG_TILING_POINTS.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_B_1of2-ICOSAHEDRON_VERTICES.pdb;
cp orig/CASOG_ONE_CAPSID_POINTS.pdb new/figs/fig5/figure_5_panel_B_2of2-CASOG_ONE_CAPSID_POINTS.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_C_1of2-ICOSAHEDRON_VERTICES.pdb;
cp orig/CASOG_TWO_CAPSID_POINTS.pdb new/figs/fig5/figure_5_panel_C_2of2-CASOG_TWO_CAPSID_POINTS.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_G_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panel_G_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
cp new/super/1TME_capsid_tiles/1TME_tile_1a.pdb new/figs/fig5/figure_5_panel_G_3of3-1TME_tile_1a.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_H_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panel_H_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
cp new/super/1TME_capsid_tiles/1TME_tile_1b.pdb new/figs/fig5/figure_5_panel_H_3of3-1TME_tile_1b.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_I_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panel_I_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
cp new/super/1TME_capsid_tiles/1TME_tile_1c.pdb new/figs/fig5/figure_5_panel_I_3of3-1TME_tile_1c.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_M_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panel_M_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
cp new/super/1TMF_capsid_tiles/1TMF_tile_1a.pdb new/figs/fig5/figure_5_panel_M_3of3-1TMF_tile_1a.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_N_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panel_N_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
cp new/super/1TMF_capsid_tiles/1TMF_tile_1b.pdb new/figs/fig5/figure_5_panel_N_3of3-1TMF_tile_1b.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panel_O_1of3-ICOSAHEDRON_VERTICES.pdb;
cp orig/TRI78.pdb new/figs/fig5/figure_5_panel_O_2of3-ICOSAHEDRON_EDGE_POINTS.pdb;
cp new/super/1TMF_capsid_tiles/1TMF_tile_1c.pdb new/figs/fig5/figure_5_panel_O_3of3-1TMF_tile_1c.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panels_P_S_1of2-ICOSAHEDRON_VERTICES.pdb;
cp new/super/1TME_tile_1.pdb new/figs/fig5/figure_5_panels_P_S_2of2-1TME_tile_1.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panels_Q_T_1of2-ICOSAHEDRON_VERTICES.pdb;
cp new/super/1TMF_tile_1.pdb new/figs/fig5/figure_5_panels_Q_T_2of2-1TMF_tile_1.pdb;
cp orig/ICOS135.pdb new/figs/fig5/figure_5_panels_R_U_1of3-ICOSAHEDRON_VERTICES.pdb;
cp new/super/1TME_tile_1.pdb new/figs/fig5/figure_5_panels_R_U_2of3-1TME_tile_1.pdb;
cp new/super/1TMF_tile_1.pdb new/figs/fig5/figure_5_panels_R_U_3of3-1TMF_tile_1.pdb;
# Figure 6. Comparison of constructed CASOG-1 and CASOG-2 picornavirus capsid tiling piece pentamers.
# (Combine aligned individual VP1234 trimers, "3xVP1234 capsid tiles".)
combinePDB --input1=orig/TRI78.pdb --input2=orig/ICOS135.pdb --nopositionchange --output=new/tmpicosframe.pdb;
cp new/tmpicosframe.pdb new/figs/fig6/figure_6_panels_A_D_G_J_1of2-ICOSAHEDRON.pdb;
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_1a.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_1b.pdb \
--output=new/tmp1.pdb \
--nopositionchange --chainid="ABCDEFGH";
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_1c.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_2a.pdb \
--output=new/tmp2.pdb \
--nopositionchange --chainid="IJKLMNOP";
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_2b.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_2c.pdb \
--output=new/tmp3.pdb \
--nopositionchange --chainid="QRSTUVWX";
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_3a.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_3b.pdb \
--output=new/tmp4.pdb \
--nopositionchange --chainid="YZ123456";
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_3c.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_4a.pdb \
--output=new/tmp5.pdb \
--nopositionchange --chainid="abcdefgh";
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_4b.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_4c.pdb \
--output=new/tmp6.pdb \
--nopositionchange --chainid="ijklmnop";
combinePDB --input1=new/super/1TME_capsid_tiles/1TME_tile_5a.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_5b.pdb \
--output=new/tmp7.pdb \
--nopositionchange --chainid="qrstuvwx";
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--output=new/tmp1234.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp5.pdb \
--input2=new/tmp6.pdb \
--output=new/tmp56.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp7.pdb \
--input2=new/super/1TME_capsid_tiles/1TME_tile_5c.pdb \
--output=new/tmp78.pdb \
--nopositionchange --chainid="qrstuvwxyz78";
combinePDB --input1=new/tmp56.pdb \
--input2=new/tmp78.pdb \
--output=new/tmp5678.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1234.pdb \
--input2=new/tmp5678.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig6/figure_6_panels_B_E_H_K_1of2-ICOSAHEDRON.pdb;
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_1a.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_1b.pdb \
--output=new/tmp1.pdb \
--nopositionchange --chainid="ABCDEFGH";
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_1c.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_2a.pdb \
--output=new/tmp2.pdb \
--nopositionchange --chainid="IJKLMNOP";
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_2b.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_2c.pdb \
--output=new/tmp3.pdb \
--nopositionchange --chainid="QRSTUVWX";
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_3a.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_3b.pdb \
--output=new/tmp4.pdb \
--nopositionchange --chainid="YZ123456";
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_3c.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_4a.pdb \
--output=new/tmp5.pdb \
--nopositionchange --chainid="abcdefgh";
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_4b.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_4c.pdb \
--output=new/tmp6.pdb \
--nopositionchange --chainid="ijklmnop";
combinePDB --input1=new/super/1TMF_capsid_tiles/1TMF_tile_5a.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_5b.pdb \
--output=new/tmp7.pdb \
--nopositionchange --chainid="qrstuvwx";
combinePDB --input1=new/tmp1.pdb \
--input2=new/tmp2.pdb \
--output=new/tmp12.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp3.pdb \
--input2=new/tmp4.pdb \
--output=new/tmp34.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp12.pdb \
--input2=new/tmp34.pdb \
--output=new/tmp1234.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp5.pdb \
--input2=new/tmp6.pdb \
--output=new/tmp56.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp7.pdb \
--input2=new/super/1TMF_capsid_tiles/1TMF_tile_5c.pdb \
--output=new/tmp78.pdb \
--nopositionchange --chainid="qrstuvwxyz78";
combinePDB --input1=new/tmp56.pdb \
--input2=new/tmp78.pdb \
--output=new/tmp5678.pdb \
--noidchange --nopositionchange;
combinePDB --input1=new/tmp1234.pdb \
--input2=new/tmp5678.pdb \
--noidchange --nopositionchange \
--output=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig6/figure_6_panels_C_F_I_L_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
new/figs/fig6/figure_6_panels_C_F_I_L_2of3-1TME_3xVP1234_PENTAMER.pdb;
cp new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
new/figs/fig6/figure_6_panels_C_F_I_L_3of3-1TMF_3xVP1234_PENTAMER.pdb;
rm new/tmp1.pdb;
rm new/tmp2.pdb;
rm new/tmp3.pdb;
rm new/tmp4.pdb;
rm new/tmp5.pdb;
rm new/tmp6.pdb;
rm new/tmp7.pdb;
rm new/tmp8.pdb;
rm new/tmp12.pdb;
rm new/tmp34.pdb;
rm new/tmp56.pdb;
rm new/tmp78.pdb;
rm new/tmp1234.pdb;
rm new/tmp5678.pdb;
# Figure 7. Construction of myelin P2 trimers, CRABP trimers, and Alpha-bungarotoxin Trimers.
#
# stripPDB is used to transform the EV-D68 (4WM7.pdb based) 3xVP1234 capsid tile to
# an EV-D68 toxin core trimer by removing chains '2': BFJ, '3': CGK, and '4': DHL,
# and by removing individual residues (1 - 78, 110 - 219) from VP1 chain '1': AEI.
#
# Note that the critical residues 81 through 86 (inclusive) are missing from 4WM7.pdb,
# and as such unfortunately do not show up in the newly created EV-D68 toxin core trimer.
#
#
# 2ABX trimer chain identifier ordinal: 1 2
# First 2ABX chain identifiers: A B
# Second 2ABX chain identifiers: C D
# Third 2ABX chain identifiers: E F
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
cp orig/TRI78.pdb new/figs/fig7/figure_7_panel_A_1of2-ICOSAHEDRON_EDGE_POINTS.pdb;
stripPDB --input=orig/PICORNAVIRUS_TILING_POINTS.pdb \
--output=new/figs/fig7/figure_7_panel_A_2of2-PICORNAVIRUS_TILING_POINTS.pdb \
--remove="1,,1;1,,4;1,,7;";
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_B_1of2-ICOSAHEDRON.pdb;
cp new/figs/fig7/figure_7_panel_A_2of2-PICORNAVIRUS_TILING_POINTS.pdb new/figs/fig7/figure_7_panel_B_2of2-PICORNAVIRUS_TILING_POINTS.pdb;
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_C_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig7/figure_7_panel_A_2of2-PICORNAVIRUS_TILING_POINTS.pdb new/figs/fig7/figure_7_panel_C_2of3-PICORNAVIRUS_TILING_POINTS.pdb;
cp new/super/2WUT_TRIMER_table5.pdb new/figs/fig7/figure_7_panel_C_3of3-2WUT_MP2_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_D_1of4-ICOSAHEDRON.pdb;
cp new/figs/fig7/figure_7_panel_A_2of2-PICORNAVIRUS_TILING_POINTS.pdb new/figs/fig7/figure_7_panel_D_2of4-PICORNAVIRUS_TILING_POINTS.pdb;
cp new/super/2WUT_TRIMER_table5.pdb new/figs/fig7/figure_7_panel_D_3of4-2WUT_MP2_TRIMER.pdb;
cp new/super/1CBS_TRIMER_table5.pdb new/figs/fig7/figure_7_panel_D_4of4-1CBS_CRABP_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_E_1of3-ICOSAHEDRON.pdb;
cp new/super/2ABX_TRIMER_table6.pdb new/figs/fig7/figure_7_panel_E_2of3-2ABX_TRIMER.pdb;
# The following atom tiling is from Table 6. "Alpha-bungarotoxin and EV-D68 VP1 trimer tiling atoms." of:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# 2ABX.pdb trimer tiling atoms are located in chains A, C, and E:
#
# Atom Type Residue Chain Atom Serial #
# --------- --------- --------- -------------
# O P10 A 69
# C 1172
# E 2275
# CA V14 A 91
# C 1194
# E 2297
# CB V14 A 94
# C 1197
# E 2300
#
# Note that the third stripPDB "triplet" field is used to specify atom serial numbers.
stripPDB --input=new/super/2ABX_TRIMER_table6.pdb \
--output=new/figs/fig7/figure_7_panel_E_3of3-2ABX_TRIMER_TILING_ATOMS.pdb \
--remove=",,-68;,,70-90;,,92-93;,,95-1171;,,1173-1193;,,1195-1196;,,1198-2274;,,2276-2296;,,2298-2299;,,2301-;";
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_F_1of3-ICOSAHEDRON.pdb;
# VP1 B-1: ABT CHAIN B
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
#
# figure_7_panel_F_2of3-4WM7_TOXIN_CORE_VP1_TRIMER.pdb is ABT CHAIN B ATOMS IN 4WM7
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-78;A,85-88;A,94-98;A,110-;B;C;D;\
E,-78;E,85-88;E,94-98;E,110-;F;G;H;\
I,-78;I,85-88;I,94-98;I,110-;J;K;L;"\
--output=new/figs/fig7/figure_7_panel_F_2of3-4WM7_TOXIN_CORE_VP1_TRIMER.pdb;
# The following atom tiling is from Table 6. "Alpha-bungarotoxin and EV-D68 VP1 trimer tiling atoms." of:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# 4WM7.pdb toxin core trimer tiling atoms are located in chains A, chains E, and I:
#
# Atom Type Residue Chain Atom Serial #
# --------- --------- --------- -------------
# CA P185 A 1355
# E 7615
# I 13875
# CB A192 A 1412
# E 7672
# I 13932
# C Y193 A 1415
# E 7675
# I 13935
#
# Note that the third stripPDB "triplet" field is used to specify atom serial numbers.
stripPDB --input=new/super/4WM7_tile_1.pdb \
--output=new/figs/fig7/figure_7_panel_F_3of3-4WM7_TOXIN_CORE_TRIMER_TILING_ATOMS.pdb \
--remove=",,-1354;,,1356-1411;,,1413-1414;,,1416-7614;,,7616-7671;,,7673-7674;,,7676-13874;,,13876-13931;,,13933-13934;,,13936-;";
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_G_1of5-ICOSAHEDRON.pdb;
cp new/super/2ABX_TRIMER_table6.pdb new/figs/fig7/figure_7_panel_G_2of5-2ABX_TRIMER.pdb;
cp new/figs/fig7/figure_7_panel_F_2of3-4WM7_TOXIN_CORE_VP1_TRIMER.pdb new/figs/fig7/figure_7_panel_G_3of5-4WM7_TOXIN_CORE_VP1_TRIMER.pdb
cp new/figs/fig7/figure_7_panel_E_3of3-2ABX_TRIMER_TILING_ATOMS.pdb new/figs/fig7/figure_7_panel_G_4of5-2ABX_TRIMER_TILING_ATOMS.pdb
cp new/figs/fig7/figure_7_panel_F_3of3-4WM7_TOXIN_CORE_TRIMER_TILING_ATOMS.pdb new/figs/fig7/figure_7_panel_G_5of5-4WM7_TOXIN_CORE_TRIMER_TILING_ATOMS.pdb
cp new/tmpicosframe.pdb new/figs/fig7/figure_7_panel_H_1of3-ICOSAHEDRON.pdb;
cp new/super/2ABX_TRIMER_table6.pdb new/figs/fig7/figure_7_panel_H_2of3-2ABX_TRIMER.pdb;
cp new/figs/fig7/figure_7_panel_F_2of3-4WM7_TOXIN_CORE_VP1_TRIMER.pdb new/figs/fig7/figure_7_panel_H_3of3-4WM7_TOXIN_CORE_VP1_TRIMER.pdb
# Figure 8. Myelin P2, CRABP, and Picornavirus Myelin P2/CRABP Cores.
#
# Atoms are stripped from Myelin/CRABP trimers to make trimer substructures showing
# Myelin P2/CRABP Cores and Helices.
#
# Atoms are stripped from VP1234 trimers to make VP1-, VP2-, VP3-, and VP123- trimer substructures
# showing regions corresponding to Myelin P2/CRABP Cores and Helices.
#
# The following is useful for panels in columns 2 - 6:
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
# Strip atoms to make Figure 8 substructures of Myelin/CRABP trimers.
#===================================================================================================
# 1CBS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# M-1: PNFSGNWKIIR [1 - 11] MP2 NON-HELIX
# M-2: SENFEELL [12 - 19] MP2 HELIX ONE
# M-3: KVLG [20 - 23] MP2 HELIX ONE
# M-4: VNVMLRKIAVAAASK [24 - 38] MP2 HELIX TWO
# M-5: PAVEIKQEGDTFYIKTSTT [39 - 57] MP2 NON-HELIX
# M-6: VRTTEINFKVG [58 - 68] MP2 NON-HELIX
# M-7: EEFEEQT [69 - 75] MP2 NON-HELIX
# M-8: VD [76 - 77] MP2 NON-HELIX
# M-9: GR [78 - 79] MP2 NON-HELIX
# M-10: PCKS [80 - 83] MP2 NON-HELIX
# M-11: LV [84 - 85] MP2 NON-HELIX
# M-12: KWESENKMVCEQKLLK [86 - 101] MP2 NON-HELIX
# M-13: GEGPKTSWT [102 - 110] MP2 NON-HELIX
# M-14: RELTNDGELILTM [111 - 123] MP2 NON-HELIX
# M-15: TADDVVCT [124 - 131] MP2 NON-HELIX
# M-16: RVYVRE [132 - 137] MP2 NON-HELIX
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A0_B0_1of4-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1CBS_TRIMER_table5.pdb --remove="\
A,12-38;\
B,12-38;\
C,12-38;"\
--output=new/figs/fig8/figure_8_panels_A0_B0_2of4-CRABP_1CBS_NON_HELIX.pdb;
stripPDB --input=new/super/1CBS_TRIMER_table5.pdb --remove="\
A,-11;A,24-;\
B,-11;B,24-;\
C,-11;C,24-;"\
--output=new/figs/fig8/figure_8_panels_A0_B0_3of4-CRABP_1CBS_HELIX_ONE.pdb;
stripPDB --input=new/super/1CBS_TRIMER_table5.pdb --remove="\
A,-23;A,39-;\
B,-23;B,39-;\
C,-23;C,39-;"\
--output=new/figs/fig8/figure_8_panels_A0_B0_4of4-CRABP_OF_1CBS_HELIX_TWO.pdb;
#===================================================================================================
# 2WUT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# M-1: GMSNKFLGTWKLVS [(-1) - 12] MP2 NON-HELIX
# M-2: SENFDDYM [13 - 20] MP2 HELIX ONE
# M-3: KALG [21 - 24] MP2 HELIX ONE
# M-4: VGLATRKLGNLAK [25 - 37] MP2 HELIX TWO
# M-5: PTVIISKKGDIITIRTEST [38 - 56] MP2 NON-HELIX
# M-6: FKNTEISFKLG [57 - 67] MP2 NON-HELIX
# M-7: QEFEETT [68 - 74] MP2 NON-HELIX
# M-8: ADN [75 - 77] MP2 NON-HELIX
# M-9: RKT [78 - 80] MP2 NON-HELIX
# M-10: KS [81 - 82] MP2 NON-HELIX
# M-11: IV [83 - 84] MP2 NON-HELIX
# M-12: TLQRGSLNQVQR [85 - 96] MP2 NON-HELIX
# M-13: WDGKETTI [97 - 104] MP2 NON-HELIX
# M-14: KRKLVNGKMVAECKMK [105 - 120] MP2 NON-HELIX
# M-15: GVVCT [121 - 125] MP2 NON-HELIX
# M-16: RIYEKV [126 - 131] MP2 NON-HELIX
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A1_B1_1of4-ICOSAHEDRON.pdb;
stripPDB --input=new/super/2WUT_TRIMER_table5.pdb --remove="\
A,13-37;\
B,13-37;\
C,13-37;"\
--output=new/figs/fig8/figure_8_panels_A1_B1_2of4-MP2_2WUT_NON_HELIX.pdb;
stripPDB --input=new/super/2WUT_TRIMER_table5.pdb --remove="\
A,-12;A,25-;\
B,-12;B,25-;\
C,-12;C,25-;"\
--output=new/figs/fig8/figure_8_panels_A1_B1_3of4-MP2_2WUT_HELIX_ONE.pdb;
stripPDB --input=new/super/2WUT_TRIMER_table5.pdb --remove="\
A,-24;A,38-;\
B,-24;B,38-;\
C,-24;C,38-;"\
--output=new/figs/fig8/figure_8_panels_A1_B1_4of4-MP2_2WUT_HELIX_TWO.pdb;
# Strip atoms to make Figure 8 VP1 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: TTSAGESADPVTTTVENYGG [1 - 20] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: ETQI [21 - 24] MP2 HELIX ONE
# VP1 I123-1: Q [25] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: RR [26 - 27] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHTDVSFIMDR [28 - 38] MP2 HELIX TWO
# VP1 M-5: FVKVTPQNQI [39 - 48] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: N [49] INSERTION
# VP1 B-1: IL [50 - 51] ABT CHAIN B
# VP1 I1-1: INSERTION
# VP1 B-2: ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: ABT CHAIN B
# VP1 I1-2: D [52] INSERTION
# VP1 B-4: LM [53 - 54] ABT CHAIN B
# VP1 I123-3: QVPSHT [55 - 60] INSERTION
# VP1 I1-3: LV [61 - 62] INSERTION
# VP1 I123-4: GG [63 - 64] INSERTION
# VP1 B-5: LLRAS [65 - 69] ABT CHAIN B
# VP1 M-6: TYYFSDLEIAVKH [70 - 82] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: EG [83 - 84] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: D [85] MP2 NON-HELIX
# VP1 I123-6: LT [86 - 87] INSERTION
# VP1 M-9: MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WV [88 - 89] MP2 NON-HELIX
# VP1 I13-2: PNGA [90 - 93] INSERTION
# VP1 I123-7: PE [94 - 95] INSERTION
# VP1 M-12: KALDNTTNPTAYHKAPLTR [96 - 114] MP2 NON-HELIX
# VP1 A-1: LALPYTAPHRVLATVYNGECRYSRNAV (PDB MISSING 'RYSRNAV') [115 - 141] ABT CHAIN A
# VP1 I1-4: PNLRGDLQVLAQ (PDB MISSING 'PNLRGDLQVLAQ') [142 - 153] INSERTION
# VP1 A-2: KVARTLPTSFNY (RES 154 - 156 'KVA' MISSING FROM PDB) [154 - 165] ABT CHAIN A
# VP1 M-13: GAIKAT [166 - 171] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: RVTELLYRMKRAETYCPRPLLA [172 - 193] MP2 NON-HELIX
# VP1 A-3: I [194] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: H [195] ABT CHAIN A
# VP1 M-15: PTEA [196 - 199] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RHKQKIVAPVK (PDB MISSING 'VK') [200 - 210] MP2 NON-HELIX
# VP1 A-6: QTL (PDB MISSING 'QTL') [211 - 213] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A2_B2_1of4-ICOSAHEDRON.pdb;
# VP1 M-1: TTSAGESADPVTTTVENYGG [1 - 20] MP2 NON-HELIX
# VP1 M-5: FVKVTPQNQI [39 - 48] MP2 NON-HELIX
# VP1 M-6: TYYFSDLEIAVKH [70 - 82] MP2 NON-HELIX
# VP1 M-7: EG [83 - 84] MP2 NON-HELIX
# VP1 M-8: D [85] MP2 NON-HELIX
# VP1 M-9: MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-11: WV [88 - 89] MP2 NON-HELIX
# VP1 M-12: KALDNTTNPTAYHKAPLTR [96 - 114] MP2 NON-HELIX
# VP1 M-13: GAIKAT [166 - 171] MP2 NON-HELIX
# VP1 M-14: RVTELLYRMKRAETYCPRPLLA [172 - 193] MP2 NON-HELIX
# VP1 M-15: PTEA [196 - 199] MP2 NON-HELIX
# VP1 M-16: RHKQKIVAPVK (PDB MISSING 'VK') [200 - 210] MP2 NON-HELIX
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,21-38;A,49-69;A,86-87;A,90-95;A,115-165;A,194-195;B;C;D;\
E,21-38;E,49-69;E,86-87;E,90-95;E,115-165;E,194-195;F;G;H;\
I,21-38;I,49-69;I,86-87;I,90-95;I,115-165;I,194-195;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A2_B2_2of4-MP2_CRABP_CORE_OF_1BBT_VP1_NON_HELIX.pdb;
# VP1 M-2: ETQI [21 - 24] MP2 HELIX ONE
# VP1 M-3: RR [26 - 27] MP2 HELIX ONE
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,-20;A,25;A,28-;B;C;D;\
E,-20;E,25;E,28-;F;G;H;\
I,-20;I,25;I,28-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A2_B2_3of4-MP2_CRABP_CORE_OF_1BBT_VP1_HELIX_ONE.pdb;
# VP1 M-4: QHTDVSFIMDR [28 - 38] MP2 HELIX TWO
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,-27;A,39-;B;C;D;\
E,-27;E,39-;F;G;H;\
I,-27;I,39-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A2_B2_4of4-MP2_CRABP_CORE_OF_1BBT_VP1_HELIX_TWO.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GVDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FNY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDYRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSSSGPQKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGVN [99 - 102] INSERTION
# VP1 I123-4: GAN [103 - 105] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [106 - 120] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 I123-6: ASVL [142 - 145] INSERTION
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [147 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGA [149 - 152] INSERTION
# VP1 I123-7: PAD [153 - 155] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [156 - 176] MP2 NON-HELIX
# VP1 A-1: LVGAGNSQVSFVVPYNSPLSVLPAAWFNGWS [177 - 207] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [209 - 220] INSERTION
# VP1 A-2: ADFGRLWI [221 - 228] ABT CHAIN A
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [251 - 256] MP2 NON-HELIX
# VP1 I2-6: PT [257 - 258] INSERTION
# VP1 M-16: PTTTKINADNPVPILELE [259 - 276] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A3_B3_1of4-ICOSAHEDRON.pdb;
# VP1 M-1: GVDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-11: WA [147 - 148] MP2 NON-HELIX
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [156 - 176] MP2 NON-HELIX
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
# VP1 M-15: TLFFPW [251 - 256] MP2 NON-HELIX
# VP1 M-16: PTTTKINADNPVPILELE [259 - 276] MP2 NON-HELIX
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,11-39;A,56-120;A,142-145;A,149-155;A,177-228;A,257-258;B;C;D;\
E,11-39;E,56-120;E,142-145;E,149-155;E,177-228;E,257-258;F;G;H;\
I,11-39;I,56-120;I,142-145;I,149-155;I,177-228;I,257-258;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A3_B3_2of4-MP2_CRABP_CORE_OF_1TMF_VP1_NON_HELIX.pdb;
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-10;A,22;A,27-;B;C;D;\
E,-10;E,22;E,27-;F;G;H;\
I,-10;I,22;I,27-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A3_B3_3of4-MP2_CRABP_CORE_OF_1TMF_VP1_HELIX_ONE.pdb;
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-28;A,40-;B;C;D;\
E,-28;E,40-;F;G;H;\
I,-28;I,40-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A3_B3_4of4-MP2_CRABP_CORE_OF_1TMF_VP1_HELIX_TWO.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GSDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNIEST [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FVY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDLRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSTSGPVKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGT [99 - 101] INSERTION
# VP1 I123-4: TN [102 - 103] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [104 - 118] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [119 - 134] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [135 - 138] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [139] MP2 NON-HELIX
# VP1 I123-6: ASVL [140 - 143] INSERTION
# VP1 M-9: R [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [145 - 146] MP2 NON-HELIX
# VP1 I13-2: PTGA [147 - 150] INSERTION
# VP1 I123-7: PAD [151 - 153] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [154 - 174] MP2 NON-HELIX
# VP1 A-1: LVGAGNTQISFVVPYNSPLSVLPAAWFNGWS [175 - 205] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [206 - 218] INSERTION
# VP1 A-2: ADFGRLWI [219 - 226] ABT CHAIN A
# VP1 M-13: QGNTSA [227 - 232] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [233 - 248] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [249 - 254] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: PVSTRSKINADNPVPILELE [255 - 274] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A4_B4_1of4-ICOSAHEDRON.pdb;
# VP1 M-1: GSDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 M-5: AVPIGMLRPGQNIEST [40 - 55] MP2 NON-HELIX
# VP1 M-6: PFTYYKCDLEVTVSAL [119 - 134] MP2 NON-HELIX
# VP1 M-7: GTDT [135 - 138] MP2 NON-HELIX
# VP1 M-8: V [139] MP2 NON-HELIX
# VP1 M-9: R [144] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-11: WA [145 - 146] MP2 NON-HELIX
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [154 - 174] MP2 NON-HELIX
# VP1 M-13: QGNTSA [227 - 232] MP2 NON-HELIX
# VP1 M-14: SVRIRYKKMKVFCPRP [233 - 248] MP2 NON-HELIX
# VP1 M-15: TLFFPW [249 - 254] MP2 NON-HELIX
# VP1 M-16: PVSTRSKINADNPVPILELE [255 - 274] MP2 NON-HELIX
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,11-39;A,56-118;A,140-143;A,147-153;A,175-226;B;C;D;\
E,11-39;E,56-118;E,140-143;E,147-153;E,175-226;F;G;H;\
I,11-39;I,56-118;I,140-143;I,147-153;I,175-226;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A4_B4_2of4-MP2_CRABP_CORE_OF_1TME_VP1_NON_HELIX.pdb;
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,-10;A,22;A,27-;B;C;D;\
E,-10;E,22;E,27-;F;G;H;\
I,-10;I,22;I,27-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A4_B4_3of4-MP2_CRABP_CORE_OF_1TME_VP1_HELIX_ONE.pdb;
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,-28;A,40-;B;C;D;\
E,-28;E,40-;F;G;H;\
I,-28;I,40-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A4_B4_4of4-MP2_CRABP_CORE_OF_1TME_VP1_HELIX_TWO.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PALTAVETGATNPLV [43 - 56] MP2 NON-HELIX
# VP1 I3-1: P [57] INSERTION
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 I123-1: Q [62] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHRSRS [68 - 73] MP2 HELIX TWO
# VP1 M-5: ESSIESFFARGACVTIMT [74 - 91] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: V [92] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: DN [93 - 94] INSERTION
# VP1 B-2: PAS [95 - 97] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: TT [98 - 99] ABT CHAIN B
# VP1 I1-2: NKDK [100 - 103] INSERTION
# VP1 B-4: LFAVW [104 - 108] ABT CHAIN B
# VP1 I123-3: K [109] INSERTION
# VP1 I1-3: IT [110 - 111] INSERTION
# VP1 I123-4: YK [112 - 113] INSERTION
# VP1 B-5: DTVQLRRKLEF [114 - 124] ABT CHAIN B
# VP1 M-6: FTYSRFDMELTFVVTA [125 - 140] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NFTETNN [141 - 147] MP2 NON-HELIX
# VP1 I123-5: GHA [148 - 150] INSERTION
# VP1 M-8: LNQ [151 - 153] MP2 NON-HELIX
# VP1 I123-6: VY [154 - 155] INSERTION
# VP1 M-9: Q [156] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: IMYV [157 - 160] MP2 NON-HELIX
# VP1 I13-2: PPGA [161 - 164] INSERTION
# VP1 I123-7: PVPE [165 - 168] INSERTION
# VP1 M-12: KWDDYTWQTSSNPSIFYTYGTAPAR [169 - 193] MP2 NON-HELIX
# VP1 A-1: ISVPYVGISNAYSHFYDGFSKV [194 - 215] ABT CHAIN A
# VP1 I1-4: PLKDQSAALGDSLYGAASLN [216 - 235] INSERTION
# VP1 A-2: DFGILAVRV [236 - 244] ABT CHAIN A
# VP1 M-13: VNDHNPT [245 - 251] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: KVTSKIRVYLKPKHIRVWCPRPPRAVA [252 - 278] MP2 NON-HELIX
# VP1 A-3: YYG [279 - 281] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: PGVDY [282 - 286] ABT CHAIN A
# VP1 I1-5: KD [287 - 288] INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: GTLTP [289 - 293] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: LSTKD [294 - 298] MP2 NON-HELIX
# VP1 A-6: LTTY [299 - 302] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A5_B5_1of4-ICOSAHEDRON.pdb;
# VP1 M-1: PALTAVETGATNPLV [43 - 56] MP2 NON-HELIX
# VP1 M-5: ESSIESFFARGACVTIMT [74 - 91] MP2 NON-HELIX
# VP1 M-6: FTYSRFDMELTFVVTA [125 - 140] MP2 NON-HELIX
# VP1 M-7: NFTETNN [141 - 147] MP2 NON-HELIX
# VP1 M-8: LNQ [151 - 153] MP2 NON-HELIX
# VP1 M-9: Q [156] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-11: IMYV [157 - 160] MP2 NON-HELIX
# VP1 M-12: KWDDYTWQTSSNPSIFYTYGTAPAR [169 - 193] MP2 NON-HELIX
# VP1 M-13: VNDHNPT [245 - 251] MP2 NON-HELIX
# VP1 M-14: KVTSKIRVYLKPKHIRVWCPRPPRAVA [252 - 278] MP2 NON-HELIX
# VP1 M-15: GTLTP [289 - 293] MP2 NON-HELIX
# VP1 M-16: LSTKD [294 - 298] MP2 NON-HELIX
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-42;A,57-73;A,92-124;A,148-150;A,154-155;A,161-168;A,194-244;A,279-288;A,299-;B;C;D;\
E,-42;E,57-73;E,92-124;E,148-150;E,154-155;E,161-168;E,194-244;E,279-288;E,299-;F;G;H;\
I,-42;I,57-73;I,92-124;I,148-150;I,154-155;I,161-168;I,194-244;I,279-288;I,299-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A5_B5_2of4-MP2_CRABP_CORE_OF_1HXS_VP1_NON_HELIX.pdb;
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-57;A,62;A,68-;B;C;D;\
E,-57;E,62;E,68-;F;G;H;\
I,-57;I,62;I,68-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A5_B5_3of4-MP2_CRABP_CORE_OF_1HXS_VP1_HELIX_ONE.pdb;
# VP1 M-4: QHRSRS [68 - 73] MP2 HELIX TWO
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-67;A,74-;B;C;D;\
E,-67;E,74-;F;G;H;\
I,-67;I,74-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A5_B5_4of4-MP2_CRABP_CORE_OF_1HXS_VP1_HELIX_TWO.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_A6_B6_1of4-ICOSAHEDRON.pdb;
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-23;A,37-55;A,74-109;A,135-138;A,141-143;A,149-155;A,183-224;A,232;A,258-273;A,291-;B;C;D;\
E,-23;E,37-55;E,74-109;E,135-138;E,141-143;E,149-155;E,183-224;E,232;E,258-273;E,291-;F;G;H;\
I,-23;I,37-55;I,74-109;I,135-138;I,141-143;I,149-155;I,183-224;I,232;I,258-273;I,291-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A6_B6_2of4-MP2_CRABP_CORE_OF_4WM7_VP1_NON_HELIX.pdb;
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-36;A,44;A,49-;B;C;D;\
E,-36;E,44;E,49-;F;G;H;\
I,-36;I,44;I,49-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A6_B6_3of4-MP2_CRABP_CORE_OF_4WM7_VP1_HELIX_ONE.pdb;
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-48;A,56-;B;C;D;\
E,-48;E,56-;F;G;H;\
I,-48;I,56-;J;K;L;"\
--output=new/figs/fig8/figure_8_panels_A6_B6_4of4-MP2_CRABP_CORE_OF_4WM7_VP1_HELIX_TWO.pdb;
# Strip atoms to make Figure 8 VP2 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: DKK (RES 1-3 'DKK' MISSING FROM PDB) [1 - 3] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: TEETTLL (RES 4-8 'TEETT' MISSING FROM PDB) [4 - 10] MP2 HELIX ONE
# VP2 I123-1: E [11] INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: D [12] INSERTION
# VP2 M-3: RI [13 - 14] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LTTRNGH [15 - 21] MP2 HELIX TWO
# VP2 M-5: TTSTTQSSVG [22 - 31] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: VTYG [32 - 35] ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: YATA [36 - 39] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: E [40] ABT CHAIN B
# VP2 I123-3: INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: INSERTION
# VP2 B-5: DFVSGP [41 - 46] ABT CHAIN B
# VP2 M-6: NTSGLETRVV [47 - 56] MP2 NON-HELIX
# VP2 I2-1: QA [57 - 58] INSERTION
# VP2 I23-2: ERFFKTHLFDW [59 - 69] INSERTION
# VP2 M-7: VTSDS [70 - 74] MP2 NON-HELIX
# VP2 I123-5: FGR [75 - 77] INSERTION
# VP2 M-8: MP2 NON-HELIX
# VP2 I123-6: CHLL [78 - 81] INSERTION
# VP2 M-9: MP2 NON-HELIX
# VP2 I23-3: E [82] INSERTION
# VP2 I2-2: INSERTION
# VP2 M-10: MP2 NON-HELIX
# VP2 I2-3: INSERTION
# VP2 M-11: L [83] MP2 NON-HELIX
# VP2 I13-2: P [84] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: TDHKGVYGSLTDSYA [85 - 99] MP2 NON-HELIX
# VP2 A-1: YMRNGWDVEVTAV [100 - 112] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: GNQF [113 - 116] ABT CHAIN A
# VP2 M-13: NGGCLL [117 - 122] MP2 NON-HELIX
# VP2 I2-4: VAM [123 - 125] INSERTION
# VP2 M-14: VPELCSIQKRE [126 - 136] MP2 NON-HELIX
# VP2 A-3: ABT CHAIN A
# VP2 I2-5: LYQLTLFPHQFINPRTNMTAHITVPFVGVNR [137 - 167] INSERTION
# VP2 A-4: YDQY [168 - 171] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: KVHK [172 - 175] ABT CHAIN A
# VP2 M-15: PWTLVVMVVA [176 - 185] MP2 NON-HELIX
# VP2 I2-6: PLTVNTEGAPQIKVYANIAPTN [186 - 207] INSERTION
# VP2 M-16: VHVAGEFPSKE [208 - 218] MP2 NON-HELIX
# VP2 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_C2_D2_1of4-ICOSAHEDRON.pdb;
# VP2 M-1: DKK (RES 1-3 'DKK' MISSING FROM PDB) [1 - 3] MP2 NON-HELIX
# VP2 M-5: TTSTTQSSVG [22 - 31] MP2 NON-HELIX
# VP2 M-6: NTSGLETRVV [47 - 56] MP2 NON-HELIX
# VP2 M-7: VTSDS [70 - 74] MP2 NON-HELIX
# VP2 M-8: MP2 NON-HELIX
# VP2 M-9: MP2 NON-HELIX
# VP2 M-10: MP2 NON-HELIX
# VP2 M-11: L [83] MP2 NON-HELIX
# VP2 M-12: TDHKGVYGSLTDSYA [85 - 99] MP2 NON-HELIX
# VP2 M-13: NGGCLL [117 - 122] MP2 NON-HELIX
# VP2 M-14: VPELCSIQKRE [126 - 136] MP2 NON-HELIX
# VP2 M-15: PWTLVVMVVA [176 - 185] MP2 NON-HELIX
# VP2 M-16: VHVAGEFPSKE [208 - 218] MP2 NON-HELIX
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
B,4-21;B,32-46;B,57-69;B,75-82;B,84;B,100-116;B,123-125;B,137-175;B,186-207;A;C;D;\
F,4-21;F,32-46;F,57-69;F,75-82;F,84;F,100-116;F,123-125;F,137-175;F,186-207;E;G;H;\
J,4-21;J,32-46;J,57-69;J,75-82;J,84;J,100-116;J,123-125;J,137-175;J,186-207;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C2_D2_2of4-MP2_CRABP_CORE_OF_1BBT_VP2_NON_HELIX.pdb;
# VP2 M-2: TEETTLL (RES 4-8 'TEETT' MISSING FROM PDB) [4 - 10] MP2 HELIX ONE
# VP2 M-3: RI [13 - 14] MP2 HELIX ONE
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
B,-3;B,11-12;B,15-;A;C;D;\
F,-3;F,11-12;F,15-;E;G;H;\
J,-3;J,11-12;J,15-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C2_D2_3of4-MP2_CRABP_CORE_OF_1BBT_VP2_HELIX_ONE.pdb;
# VP2 M-4: LTTRNGH [15 - 21] MP2 HELIX TWO
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
B,-14;B,22-;A;C;D;\
F,-14;F,22-;E;G;H;\
J,-14;J,22-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C2_D2_4of4-MP2_CRABP_CORE_OF_1BBT_VP2_HELIX_TWO.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: DQNTEEM [1 - 7] MP2 HELIX ONE
# VP2 I123-1: E [8] INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: NLSD [9 - 12] INSERTION
# VP2 M-3: RVA [13 - 15] MP2 HELIX ONE
# VP2 I123-2: SD [16 - 17] INSERTION
# VP2 M-4: KAG [18 - 20] MP2 HELIX TWO
# VP2 M-5: NSATNTQSTVGR [21 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: L [33] ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [34] ABT CHAIN B
# VP2 I3-4: G [35] INSERTION
# VP2 B-3: Y [36] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GK [37 - 38] ABT CHAIN B
# VP2 I123-3: S [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: HHG [40 - 42] INSERTION
# VP2 B-5: EHPASCA [43 - 49] ABT CHAIN B
# VP2 M-6: DTATDKVL [50 - 57] MP2 NON-HELIX
# VP2 I2-1: AA [58 - 59] INSERTION
# VP2 I23-2: ERYYTIDLASWTTS [60 - 73] INSERTION
# VP2 M-7: QEAFS [74 - 78] MP2 NON-HELIX
# VP2 I123-5: HIRIPLP [79 - 85] INSERTION
# VP2 M-8: MP2 NON-HELIX
# VP2 I123-6: HVLAG [86 - 90] INSERTION
# VP2 M-9: MP2 NON-HELIX
# VP2 I23-3: EDGG [91 - 94] INSERTION
# VP2 I2-2: VFGATLRRHYL [95 - 105] INSERTION
# VP2 M-10: CKTG [106 - 109] MP2 NON-HELIX
# VP2 I2-3: WRVQVQCNASQFHAGSLL [110 - 127] INSERTION
# VP2 M-11: VFMAP [128 - 132] MP2 NON-HELIX
# VP2 I13-2: INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EFYTGKGTKTGTMEP [133 - 147] MP2 NON-HELIX
# VP2 A-1: SDPFTMDTEWRS P [148 - 160] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: QGAPTGYRY [161 - 169] ABT CHAIN A
# VP2 M-13: DSRTGFF [170 - 176] MP2 NON-HELIX
# VP2 I2-4: ATN [177 - 179] INSERTION
# VP2 M-14: HQNQW [180 - 184] MP2 NON-HELIX
# VP2 A-3: ABT CHAIN A
# VP2 I2-5: QWTVYPHQILNLRTNTTVDLEVPYVNVAP [185 - 213] INSERTION
# VP2 A-4: SSSWTQ [214 - 219] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: H [220] ABT CHAIN A
# VP2 M-15: ANWTLVVAVLS [221 - 231] MP2 NON-HELIX
# VP2 I2-6: PLQYATGSSPDVQITASLQPVNPVFNG [232 - 258] INSERTION
# VP2 M-16: LRHETV [259 - 264] MP2 NON-HELIX
# VP2 A-6: IAQ [265 - 267] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_C3_D3_1of4-ICOSAHEDRON.pdb;
# VP2 M-1: MP2 NON-HELIX
# VP2 M-5: NSATNTQSTVGR [21 - 32] MP2 NON-HELIX
# VP2 M-6: DTATDKVL [50 - 57] MP2 NON-HELIX
# VP2 M-7: QEAFS [74 - 78] MP2 NON-HELIX
# VP2 M-8: MP2 NON-HELIX
# VP2 M-9: MP2 NON-HELIX
# VP2 M-10: CKTG [106 - 109] MP2 NON-HELIX
# VP2 M-11: VFMAP [128 - 132] MP2 NON-HELIX
# VP2 M-12: EFYTGKGTKTGTMEP [133 - 147] MP2 NON-HELIX
# VP2 M-13: DSRTGFF [170 - 176] MP2 NON-HELIX
# VP2 M-14: HQNQW [180 - 184] MP2 NON-HELIX
# VP2 M-15: ANWTLVVAVLS [221 - 231] MP2 NON-HELIX
# VP2 M-16: LRHETV [259 - 264] MP2 NON-HELIX
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
B,-20;B,33-49;B,58-73;B,79-105;B,110-127;B,148-169;B,177-179;B,185-220;B,232-258;B,265-;A;C;D;\
F,-20;F,33-49;F,58-73;F,79-105;F,110-127;F,148-169;F,177-179;F,185-220;F,232-258;F,265-;E;G;H;\
J,-20;J,33-49;J,58-73;J,79-105;J,110-127;J,148-168;J,177-179;J,185-220;J,232-258;J,265-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C3_D3_2of4-MP2_CRABP_CORE_OF_1TMF_VP2_NON_HELIX.pdb;
# VP2 M-2: DQNTEEM [1 - 7] MP2 HELIX ONE
# VP2 M-3: RVA [13 - 15] MP2 HELIX ONE
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
B,8-12;B,16-;A;C;D;\
F,8-12;F,16-;E;G;H;\
J,8-12;J,16-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C3_D3_3of4-MP2_CRABP_CORE_OF_1TMF_VP2_HELIX_ONE.pdb;
# VP2 M-4: KAG [18 - 20] MP2 HELIX TWO
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
B,-17;B,21-;A;C;D;\
F,-17;F,21-;E;G;H;\
J,-17;J,21-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C3_D3_4of4-MP2_CRABP_CORE_OF_1TMF_VP2_HELIX_TWO.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: DQNTEEM (RES 1 - 7 'DQNTTEM' MISSING FROM PDB) [1 - 7] MP2 HELIX ONE
# VP2 I123-1: E (RES 8 'E' MISSING FROM PDB) [8] INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: NLSD (RES 9 - 12 'NLS' MISSING FROM PDB) [9 - 12] INSERTION
# VP2 M-3: RVA [13 - 15] MP2 HELIX ONE
# VP2 I123-2: SD [16 - 17] INSERTION
# VP2 M-4: KAG [18 - 20] MP2 HELIX TWO
# VP2 M-5: NSATNTQSTVGR [21 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: L [33] ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [34] ABT CHAIN B
# VP2 I3-4: G [35] INSERTION
# VP2 B-3: Y [36] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GEA [37 - 39] ABT CHAIN B
# VP2 I123-3: INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: HHG [40 - 42] INSERTION
# VP2 B-5: EHPASCA [43 - 49] ABT CHAIN B
# VP2 M-6: DTATDKVL [50 - 57] MP2 NON-HELIX
# VP2 I2-1: AA [58 - 59] INSERTION
# VP2 I23-2: ERYYTIDLASWTTT [60 - 73] INSERTION
# VP2 M-7: QEAFS [74 - 78] MP2 NON-HELIX
# VP2 I123-5: HIRIPLP [79 - 85] INSERTION
# VP2 M-8: MP2 NON-HELIX
# VP2 I123-6: HVLAG [86 - 90] INSERTION
# VP2 M-9: MP2 NON-HELIX
# VP2 I23-3: EDGG [91 - 94] INSERTION
# VP2 I2-2: VFGATLRRHYL [95 - 105] INSERTION
# VP2 M-10: CKTG [106 - 109] MP2 NON-HELIX
# VP2 I2-3: WRVQVQCNASQFHAGSLL [110 - 127] INSERTION
# VP2 M-11: VFMAP [128 - 132] MP2 NON-HELIX
# VP2 I13-2: INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EFYTGKGTKTGDMEP [133 - 147] MP2 NON-HELIX
# VP2 A-1: TDPFTMDTTWRAP [148 - 160] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: QGAPTGYRY [161 - 169] ABT CHAIN A
# VP2 M-13: DSRTGFF [170 - 176] MP2 NON-HELIX
# VP2 I2-4: AMN [177 - 179] INSERTION
# VP2 M-14: HQNQW [180 - 184] MP2 NON-HELIX
# VP2 A-3: ABT CHAIN A
# VP2 I2-5: QWTVYPHQILNLRTNTTVDLEVPYVNIAP [185 - 213] INSERTION
# VP2 A-4: TSSWTQ [214 - 219] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: H [220] ABT CHAIN A
# VP2 M-15: ANWTLVVAVFS [221 - 231] MP2 NON-HELIX
# VP2 I2-6: PLQYASGSSSDVQITASIQPVNPVFNG [232 - 258] INSERTION
# VP2 M-16: LRHETV [259 - 264] MP2 NON-HELIX
# VP2 A-6: IAQ (RES 267 'Q' MISSING FROM PDB) [265 - 267] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_C4_D4_1of4-ICOSAHEDRON.pdb;
# VP2 M-1: MP2 NON-HELIX
# VP2 M-5: NSATNTQSTVGR [21 - 32] MP2 NON-HELIX
# VP2 M-6: DTATDKVL [50 - 57] MP2 NON-HELIX
# VP2 M-7: QEAFS [74 - 78] MP2 NON-HELIX
# VP2 M-8: MP2 NON-HELIX
# VP2 M-9: MP2 NON-HELIX
# VP2 M-10: CKTG [106 - 109] MP2 NON-HELIX
# VP2 M-11: VFMAP [128 - 132] MP2 NON-HELIX
# VP2 M-12: EFYTGKGTKTGDMEP [133 - 147] MP2 NON-HELIX
# VP2 M-13: DSRTGFF [170 - 176] MP2 NON-HELIX
# VP2 M-14: HQNQW [180 - 184] MP2 NON-HELIX
# VP2 M-15: ANWTLVVAVFS [221 - 231] MP2 NON-HELIX
# VP2 M-16: LRHETV [259 - 264] MP2 NON-HELIX
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
B,-20;B,33-49;B,58-73;B,79-105;B,110-127;B,148-169;B,177-179;B,185-220;B,232-258;B,265-;A;C;D;\
F,-20;F,33-49;F,58-73;F,79-105;F,110-127;F,148-169;F,177-179;F,185-220;F,232-258;F,265-;E;G;H;\
J,-20;J,33-49;J,58-73;J,79-105;J,110-127;J,148-169;J,177-179;J,185-220;J,232-258;J,265-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C4_D4_2of4-MP2_CRABP_CORE_OF_1TME_VP2_NON_HELIX.pdb;
# VP2 M-2: DQNTEEM (RES 1 - 7 'DQNTTEM' MISSING FROM PDB) [1 - 7] MP2 HELIX ONE
# VP2 M-3: RVA [13 - 15] MP2 HELIX ONE
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
B,8-12;B,16-;A;C;D;\
F,8-12;F,16-;E;G;H;\
J,8-12;J,16-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C4_D4_3of4-MP2_CRABP_CORE_OF_1TME_VP2_HELIX_ONE.pdb;
# VP2 M-4: KAG [18 - 20] MP2 HELIX TWO
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
B,-17;B,21-;A;C;D;\
F,-17;F,21-;E;G;H;\
J,-17;J,21-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C4_D4_4of4-MP2_CRABP_CORE_OF_1TME_VP2_HELIX_TWO.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: S (RES 1 'S' MISSING) [1] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: PNIEA (RES 2-5 'PNIE' MISSING) [2 - 6] MP2 HELIX ONE
# VP2 I123-1: INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: CGYSD [7 - 11] INSERTION
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LQLTLG [14 - 19] MP2 HELIX TWO
# VP2 M-5: NSTITTQEAANSV [20 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: V [33] ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: AY [34 - 35] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GRW [36 - 37] ABT CHAIN B
# VP2 I123-3: P [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: INSERTION
# VP2 B-5: EYLRDS [40 - 45] ABT CHAIN B
# VP2 M-6: EANPVDQ [46 - 52] MP2 NON-HELIX
# VP2 I2-1: PTEPDVAAC [53 - 61] INSERTION
# VP2 I23-2: RFYTLDTVSWT [62 - 72] INSERTION
# VP2 M-7: KES [73 - 75] MP2 NON-HELIX
# VP2 I123-5: RGWWWKLP [76 - 83] INSERTION
# VP2 M-8: D [84] MP2 NON-HELIX
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 M-9: R [87] MP2 NON-HELIX
# VP2 I23-3: DM [88 - 89] INSERTION
# VP2 I2-2: GLFGQNMYYHYLG [90 - 102] INSERTION
# VP2 M-10: RS [103 - 104] MP2 NON-HELIX
# VP2 I2-3: GYTVHVQCNASKFHQGALG [105 - 123] INSERTION
# VP2 M-11: VFAV [124 - 127] MP2 NON-HELIX
# VP2 I13-2: P [128] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EMCLAGDSNT [129 - 138] MP2 NON-HELIX
# VP2 A-1: TTMHTSYQNANPGEKGG [139 - 155] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: TFT [156 - 158] ABT CHAIN A
# VP2 M-13: GTFT [159 - 162] MP2 NON-HELIX
# VP2 I2-4: PDNN [163 - 166] INSERTION
# VP2 M-14: QTSPARRFCPVDYLL [167 - 181] MP2 NON-HELIX
# VP2 A-3: GNGT [182 - 185] ABT CHAIN A
# VP2 I2-5: LLGNAFVFPHQIINLRTNNCATLVLPYVNSLS [186 - 217] INSERTION
# VP2 A-4: IDSMV [218 - 222] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: KH [223 - 224] ABT CHAIN A
# VP2 M-15: NNWGIAILPLA [225 - 235] MP2 NON-HELIX
# VP2 I2-6: PLNFASESSPEIPITLTIAPMCCEFNG [236 - 263] INSERTION
# VP2 M-16: LRNITLP [263 - 269] MP2 NON-HELIX
# VP2 A-6: RLQ [270 - 272] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_C5_D5_1of4-ICOSAHEDRON.pdb;
# VP2 M-1: S (RES 1 'S' MISSING) [1] MP2 NON-HELIX
# VP2 M-5: NSTITTQEAANSV [20 - 32] MP2 NON-HELIX
# VP2 M-6: EANPVDQ [46 - 52] MP2 NON-HELIX
# VP2 M-7: KES [73 - 75] MP2 NON-HELIX
# VP2 M-8: D [84] MP2 NON-HELIX
# VP2 M-9: R [87] MP2 NON-HELIX
# VP2 M-10: RS [103 - 104] MP2 NON-HELIX
# VP2 M-11: VFAV [124 - 127] MP2 NON-HELIX
# VP2 M-12: EMCLAGDSNT [129 - 138] MP2 NON-HELIX
# VP2 M-13: GTFT [159 - 162] MP2 NON-HELIX
# VP2 M-14: QTSPARRFCPVDYLL [167 - 181] MP2 NON-HELIX
# VP2 M-15: NNWGIAILPLA [225 - 235] MP2 NON-HELIX
# VP2 M-16: LRNITLP [263 - 269] MP2 NON-HELIX
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
B,2-19;B,33-45;B,53-72;B,76-83;B,85-86;B,88-102;B,105-123;B,128;B,139-158;B,163-166;B,182-224;B,236-262;B,270-;A;C;D;\
F,2-19;F,33-45;F,53-72;F,76-83;F,85-86;F,88-102;F,105-123;F,128;F,139-158;F,163-166;F,182-224;F,236-262;F,270-;E;G;H;\
J,2-19;J,33-45;J,53-72;J,76-83;J,85-86;J,88-102;J,105-123;J,128;J,139-158;J,163-166;J,182-224;J,236-262;J,270-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C5_D5_2of4-MP2_CRABP_CORE_OF_1HXS_VP2_NON_HELIX.pdb;
# VP2 M-2: PNIEA (RES 2-5 'PNIE' MISSING) [2 - 6] MP2 HELIX ONE
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
B,1;B,7-11;B,14-;A;C;D;\
F,1;F,7-11;F,14-;E;G;H;\
J,1;J,7-11;J,14-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C5_D5_3of4-MP2_CRABP_CORE_OF_1HXS_VP2_HELIX_ONE.pdb;
# VP2 M-4: LQLTLG [14 - 19] MP2 HELIX TWO
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
B,-13;B,20-;A;C;D;\
F,-13;F,20-;E;G;H;\
J,-13;J,20-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C5_D5_4of4-MP2_CRABP_CORE_OF_1HXS_VP2_HELIX_TWO.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: S (RES 1 'S' MISSING FROM PDB) [1] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: PSAEA (RES 2-6 'PSAEA' MISSING FROM PDB) [2 - 6] MP2 HELIX ONE
# VP2 I123-1: INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: CGYSD (RES 7-9 'CGY' MISSING FROM PDB) [7 - 11] INSERTION
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LQLKLG [14 - 19] MP2 HELIX TWO
# VP2 M-5: NSAIVTQEAANYC [20 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [33] ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: AY [34 - 35] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GEW [36 - 38] ABT CHAIN B
# VP2 I123-3: P [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: N [40] INSERTION
# VP2 B-5: YLPD [41 - 44] ABT CHAIN B
# VP2 M-6: HEAVAIDK [45 - 52] MP2 NON-HELIX
# VP2 I2-1: PTQPETST [53 - 60] INSERTION
# VP2 I23-2: DRFYTLRSVKW [61 - 71] INSERTION
# VP2 M-7: ESNST [72 - 76] MP2 NON-HELIX
# VP2 I123-5: GWWWKL [77 - 82] INSERTION
# VP2 M-8: PD [83 - 84] MP2 NON-HELIX
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 M-9: NN [87 - 88] MP2 NON-HELIX
# VP2 I23-3: IGMFGQNVQYHYLY [89 - 102] INSERTION
# VP2 I2-2: RS [103 - 104] INSERTION
# VP2 M-10: GFLIHVQCNATKFHQGALL [105 - 123] MP2 NON-HELIX
# VP2 I2-3: INSERTION
# VP2 M-11: VVAI [124 - 127] MP2 NON-HELIX
# VP2 I13-2: P [128] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EHQRGAHDTT [129 - 138] MP2 NON-HELIX
# VP2 A-1: TSPGF [139 - 143] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: NDIMK [144 - 148] ABT CHAIN A
# VP2 M-13: GERGGTF [149 - 155] MP2 NON-HELIX
# VP2 I2-4: N [156] INSERTION
# VP2 M-14: HPYVL [157 - 161] MP2 NON-HELIX
# VP2 A-3: DDGTS [162 - 166] ABT CHAIN A
# VP2 I2-5: IACATIFPHQWINLRTNNSATIVLPWMNVAP [167 - 197] INSERTION
# VP2 A-4: MDFPL [198 - 202] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: RH [203 - 204] ABT CHAIN A
# VP2 M-15: NQWTLAVIPVV [205 - 215] MP2 NON-HELIX
# VP2 I2-6: PLGTRTMSSVVPITVSIAPMCCEFNG [216 - 241] INSERTION
# VP2 M-16: LRHAIT [242 - 247] MP2 NON-HELIX
# VP2 A-6: Q [248] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_C6_D6_1of4-ICOSAHEDRON.pdb;
# VP2 M-1: S (RES 1 'S' MISSING FROM PDB) [1] MP2 NON-HELIX
# VP2 M-5: NSAIVTQEAANYC [20 - 32] MP2 NON-HELIX
# VP2 M-6: HEAVAIDK [45 - 52] MP2 NON-HELIX
# VP2 M-7: ESNST [72 - 76] MP2 NON-HELIX
# VP2 M-8: PD [83 - 84] MP2 NON-HELIX
# VP2 M-9: NN [87 - 88] MP2 NON-HELIX
# VP2 M-10: GFLIHVQCNATKFHQGALL [105 - 123] MP2 NON-HELIX
# VP2 M-11: VVAI [124 - 127] MP2 NON-HELIX
# VP2 M-12: EHQRGAHDTT [129 - 138] MP2 NON-HELIX
# VP2 M-13: GERGGTF [149 - 155] MP2 NON-HELIX
# VP2 M-14: HPYVL [157 - 161] MP2 NON-HELIX
# VP2 M-15: NQWTLAVIPVV [205 - 215] MP2 NON-HELIX
# VP2 M-16: LRHAIT [242 - 247] MP2 NON-HELIX
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
B,2-19;B,33-44;B,53-71;B,77-82;B,85-86;B,89-104;B,128;B,139-148;B,156;B,162-204;B,216-241;B,248;A;C;D;\
F,2-19;F,33-44;F,53-71;F,77-82;F,85-86;F,89-104;F,128;F,139-148;F,156;F,162-204;F,216-241;F,248;E;G;H;\
J,2-19;J,33-44;J,53-71;J,77-82;J,85-86;J,89-104;J,128;J,139-148;J,156;J,162-204;J,216-241;J,248;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C6_D6_2of4-MP2_CRABP_CORE_OF_4WM7_VP2_NON_HELIX.pdb;
# VP2 M-2: PSAEA (RES 2-6 'PSAEA' MISSING FROM PDB) [2 - 6] MP2 HELIX ONE
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
B,1;B,7-11;B,14-;A;C;D;\
F,1;F,7-11;F,14-;E;G;H;\
J,1;J,7-11;J,14-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C6_D6_3of4-MP2_CRABP_CORE_OF_4WM7_VP2_HELIX_ONE.pdb;
# VP2 M-4: LQLKLG [14 - 19] MP2 HELIX TWO
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
B,1-13;B,20-;A;C;D;\
F,1-13;F,20-;E;G;H;\
J,1-13;J,20-;I;K;L;"\
--output=new/figs/fig8/figure_8_panels_C6_D6_4of4-MP2_CRABP_CORE_OF_4WM7_VP2_HELIX_TWO.pdb;
# Strip atoms to make Figure 8 VP3 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: ACPTFLRFEGGV [50 - 61] MP2 NON-HELIX
# VP3 I3-1: PYVTTK [62 - 67] INSERTION
# VP3 M-2: TDSD [68 - 71] MP2 HELIX ONE
# VP3 I123-1: INSERTION
# VP3 I3-2: RV [72 - 73] INSERTION
# VP3 I23-1: LAQFD [74 - 78] INSERTION
# VP3 M-3: MP2 HELIX ONE
# VP3 I123-2: MS [79 - 80] INSERTION
# VP3 M-4: LAAKHMSNTFLAG [81 - 93] MP2 HELIX TWO
# VP3 M-5: LAQYYT [94 - 99] MP2 NON-HELIX
# VP3 I3-3: QYS [100 - 102] INSERTION
# VP3 I13-1: GTIN [103 - 106] INSERTION
# VP3 B-1: L [107] ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: HF [108 - 109] ABT CHAIN B
# VP3 I3-4: MFTGPTDAKARYMV [110 - 123] INSERTION
# VP3 B-3: AYA [124 - 126] ABT CHAIN B
# VP3 I1-2: P [127] INSERTION
# VP3 B-4: PGME [128 - 131] ABT CHAIN B
# VP3 I123-3: P [132] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: PKT [133 - 135] INSERTION
# VP3 B-5: PEAAAHCI [136 - 143] ABT CHAIN B
# VP3 M-6: HAEWDTGLNSKFTFSIPYLSAA [144 - 165] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: DYTYTAS [166 - 172] INSERTION
# VP3 M-7: DVAETT [173 - 178] MP2 NON-HELIX
# VP3 I123-5: NVQGWVCLF [179 - 187] INSERTION
# VP3 M-8: Q [188] MP2 NON-HELIX
# VP3 I123-6: INSERTION
# VP3 M-9: MP2 NON-HELIX
# VP3 I23-3: INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: I [189] MP2 NON-HELIX
# VP3 I13-2: INSERTION
# VP3 I123-7: TH [190 - 191] INSERTION
# VP3 M-12: GKADGDALVVLASAGK [192 - 207] MP2 NON-HELIX
# VP3 A-1: DFELRLPV [208 - 215] ABT CHAIN A
# VP3 I1-4: DAR [216 - 218] INSERTION
# VP3 A-2: ABT CHAIN A
# VP3 M-13: AE [219 - 220] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_E2_F2_1of4-ICOSAHEDRON.pdb;
# VP3 M-1: ACPTFLRFEGGV [50 - 61] MP2 NON-HELIX
# VP3 M-5: LAQYYT [94 - 99] MP2 NON-HELIX
# VP3 M-6: HAEWDTGLNSKFTFSIPYLSAA [144 - 165] MP2 NON-HELIX
# VP3 M-7: DVAETT [173 - 178] MP2 NON-HELIX
# VP3 M-8: Q [188] MP2 NON-HELIX
# VP3 M-9: MP2 NON-HELIX
# VP3 M-10: MP2 NON-HELIX
# VP3 M-11: I [189] MP2 NON-HELIX
# VP3 M-12: GKADGDALVVLASAGK [192 - 207] MP2 NON-HELIX
# VP3 M-13: AE [219 - 220] MP2 NON-HELIX
# VP3 M-14: MP2 NON-HELIX
# VP3 M-15: MP2 NON-HELIX
# VP3 M-16: MP2 NON-HELIX
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
C,-49;C,62-93;C,100-143;C,166-172;C,179-187;C,190-191;C,208-218;A;B;D;\
G,-49;G,62-93;G,100-143;G,166-172;G,179-187;G,190-191;G,208-218;E;F;H;\
K,-49;K,62-93;K,100-143;K,166-172;K,179-187;K,190-191;K,208-218;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E2_F2_2of4-MP2_CRABP_CORE_OF_1BBT_VP3_NON_HELIX.pdb;
# VP3 M-2: TDSD [68 - 71] MP2 HELIX ONE
# VP3 M-3: MP2 HELIX ONE
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
C,-67;C,72-;A;B;D;\
G,-67;G,72-;E;F;H;\
K,-67;K,72-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E2_F2_3of4-MP2_CRABP_CORE_OF_1BBT_VP3_HELIX_ONE.pdb;
# VP3 M-4: LAAKHMSNTFLAG [81 - 93] MP2 HELIX TWO
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
C,-80;C,94-;A;B;D;\
G,-80;G,94-;E;F;H;\
K,-80;K,94-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E2_F2_4of4-MP2_CRABP_CORE_OF_1BBT_VP3_HELIX_TWO.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: PNTNNKRY [57 - 64] MP2 NON-HELIX
# VP3 I3-1: PYFSATNSV [65 - 73] INSERTION
# VP3 M-2: PATSMVDY [74 - 81] MP2 HELIX ONE
# VP3 I123-1: Q [82] INSERTION
# VP3 I3-2: VALSCS [83 - 88] INSERTION
# VP3 I23-1: CMAN [89 - 92] INSERTION
# VP3 M-3: S [93] MP2 HELIX ONE
# VP3 I123-2: M [94] INSERTION
# VP3 M-4: LAAVARN [95 - 101] MP2 HELIX TWO
# VP3 M-5: FNQYR [102 - 106] MP2 NON-HELIX
# VP3 I3-3: INSERTION
# VP3 I13-1: GSLN [107 - 110] INSERTION
# VP3 B-1: FL [111 - 112] ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: F [113] ABT CHAIN B
# VP3 I3-4: VFTGAAMVKGKFLI [114 - 127] INSERTION
# VP3 B-3: AYT [128 - 130] ABT CHAIN B
# VP3 I1-2: P [131] INSERTION
# VP3 B-4: PGAGK [132 - 136] ABT CHAIN B
# VP3 I123-3: P [137] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: T [138] INSERTION
# VP3 B-5: TRDQAM [139 - 144] ABT CHAIN B
# VP3 M-6: QSTYAIWDLGLNSSFNFTAPFISPTH [145 - 170] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRQTSYTSPTITS [171 - 183] INSERTION
# VP3 M-7: VD [184 - 185] MP2 NON-HELIX
# VP3 I123-5: GWVTVW [186 - 191] INSERTION
# VP3 M-8: Q [192] MP2 NON-HELIX
# VP3 I123-6: LT [193 - 194] INSERTION
# VP3 M-9: P [195] MP2 NON-HELIX
# VP3 I23-3: LT [196 - 197] INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: Y [198] MP2 NON-HELIX
# VP3 I13-2: PSG [199 - 201] INSERTION
# VP3 I123-7: TP [202 - 203] INSERTION
# VP3 M-12: TNSDILTLVSAG [204 - 215] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: DDFTLRMPISPTKWV [216 - 230] ABT CHAIN A
# VP3 M-13: P [231] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: Q [232] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_E3_F3_1of4-ICOSAHEDRON.pdb;
# VP3 M-1: PNTNNKRY [57 - 64] MP2 NON-HELIX
# VP3 M-5: FNQYR [102 - 106] MP2 NON-HELIX
# VP3 M-6: QSTYAIWDLGLNSSFNFTAPFISPTH [145 - 170] MP2 NON-HELIX
# VP3 M-7: VD [184 - 185] MP2 NON-HELIX
# VP3 M-8: Q [192] MP2 NON-HELIX
# VP3 M-9: P [195] MP2 NON-HELIX
# VP3 M-10: MP2 NON-HELIX
# VP3 M-11: Y [198] MP2 NON-HELIX
# VP3 M-12: TNSDILTLVSAG [204 - 215] MP2 NON-HELIX
# VP3 M-13: P [231] MP2 NON-HELIX
# VP3 M-14: Q [232] MP2 NON-HELIX
# VP3 M-15: MP2 NON-HELIX
# VP3 M-16: MP2 NON-HELIX
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
C,-56;C,65-101;C,107-144;C,171-183;C,186-191;C,193-194;C,196-197;C,199-203;C,216-230;A;B;D;\
G,-56;G,65-101;G,107-144;G,171-183;G,186-191;G,193-194;G,196-197;G,199-203;G,216-230;E;F;H;\
K,-56;K,65-101;K,107-144;K,171-183;K,186-191;K,193-194;K,196-197;K,199-203;K,216-230;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E3_F3_2of4-MP2_CRABP_CORE_OF_1TMF_VP3_NON_HELIX.pdb;
# VP3 M-2: PATSMVDY [74 - 81] MP2 HELIX ONE
# VP3 M-3: S [93] MP2 HELIX ONE
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
C,-73;C,82-92;C,94-;A;B;D;\
G,-73;G,82-92;G,94-;E;F;H;\
K,-73;K,82-92;K,94-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E3_F3_3of4-MP2_CRABP_CORE_OF_1TMF_VP3_HELIX_ONE.pdb;
# VP3 M-4: LAAVARN [95 - 101] MP2 HELIX TWO
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
C,-94;C,102-;A;B;D;\
G,-94;G,102-;E;F;H;\
K,-94;K,102-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E3_F3_4of4-MP2_CRABP_CORE_OF_1TMF_VP3_HELIX_TWO.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: PNSNNKRY [57 - 64] MP2 NON-HELIX
# VP3 I3-1: PYFSATNSV [65 - 73] INSERTION
# VP3 M-2: PTTSLVDY [74 - 81] MP2 HELIX ONE
# VP3 I123-1: Q [82] INSERTION
# VP3 I3-2: VALSCS [83 - 88] INSERTION
# VP3 I23-1: CMCN [89 - 92] INSERTION
# VP3 M-3: S [93] MP2 HELIX ONE
# VP3 I123-2: M [94] INSERTION
# VP3 M-4: LAAVARN [95 - 101] MP2 HELIX TWO
# VP3 M-5: FNQYR [102 - 106] MP2 NON-HELIX
# VP3 I3-3: INSERTION
# VP3 I13-1: GSLN [107 - 110] INSERTION
# VP3 B-1: FL [111 - 112] ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: F [113] ABT CHAIN B
# VP3 I3-4: VFTGAAMVKGKFLI [114 - 127] INSERTION
# VP3 B-3: AYT [128 - 130] ABT CHAIN B
# VP3 I1-2: P [131] INSERTION
# VP3 B-4: PGAGK [132 - 136] ABT CHAIN B
# VP3 I123-3: P [137] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: T [138] INSERTION
# VP3 B-5: TRDQAM [139 - 144] ABT CHAIN B
# VP3 M-6: QATYAIWDLGLNSSFVFTAPFISPTH [145 - 170] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRQTSYTSATIAS (RES 180-181 'TI' MISSING FROM PDB) [171 - 183] INSERTION
# VP3 M-7: VD [184 - 185] MP2 NON-HELIX
# VP3 I123-5: GWVTVW [186 - 191] INSERTION
# VP3 M-8: Q [192] MP2 NON-HELIX
# VP3 I123-6: LT [193 - 194] INSERTION
# VP3 M-9: P [195] MP2 NON-HELIX
# VP3 I23-3: LT [196 - 197] INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: Y [198] MP2 NON-HELIX
# VP3 I13-2: PSG [199 - 201] INSERTION
# VP3 I123-7: TPV [202 - 204] INSERTION
# VP3 M-12: NSDILTLVSAG [205 - 215] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: DDFTLRMPISPTKWV [216 - 230] ABT CHAIN A
# VP3 M-13: PQ [231 - 232] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: GSDN (RES 233-236 'GSDN' MISSING FROM PDB) [233 - 236] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_E4_F4_1of4-ICOSAHEDRON.pdb;
# VP3 M-1: PNSNNKRY [57 - 64] MP2 NON-HELIX
# VP3 M-5: FNQYR [102 - 106] MP2 NON-HELIX
# VP3 M-6: QATYAIWDLGLNSSFVFTAPFISPTH [145 - 170] MP2 NON-HELIX
# VP3 M-7: VD [184 - 185] MP2 NON-HELIX
# VP3 M-8: Q [192] MP2 NON-HELIX
# VP3 M-9: P [195] MP2 NON-HELIX
# VP3 M-10: MP2 NON-HELIX
# VP3 M-11: Y [198] MP2 NON-HELIX
# VP3 M-12: NSDILTLVSAG [205 - 215] MP2 NON-HELIX
# VP3 M-13: PQ [231 - 232] MP2 NON-HELIX
# VP3 M-14: GSDN (RES 233-236 'GSDN' MISSING FROM PDB) [233 - 236] MP2 NON-HELIX
# VP3 M-15: MP2 NON-HELIX
# VP3 M-16: MP2 NON-HELIX
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
C,-56;C,65-101;C,107-144;C,171-183;C,186-191;C,193-194;C,196-197;C,199-204;C,216-230;A;B;D;\
G,-56;G,65-101;G,107-144;G,171-183;G,186-191;G,193-194;G,196-197;G,199-204;G,216-230;E;F;H;\
K,-56;K,65-101;K,107-144;K,171-183;K,186-191;K,193-194;K,196-197;K,199-204;K,216-230;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E4_F4_2of4-MP2_CRABP_CORE_OF_1TME_VP3_NON_HELIX.pdb;
# VP3 M-2: PTTSLVDY [74 - 81] MP2 HELIX ONE
# VP3 M-3: S [93] MP2 HELIX ONE
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
C,-73;C,82-92;C,94-;A;B;D;\
G,-73;G,82-92;G,94-;E;F;H;\
K,-73;K,82-92;K,94-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E4_F4_3of4-MP2_CRABP_CORE_OF_1TME_VP3_HELIX_ONE.pdb;
# VP3 M-4: LAAVARN [95 - 101] MP2 HELIX TWO
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
C,-94;C,102-;A;B;D;\
G,-94;G,102-;E;F;H;\
K,-94;K,102-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E4_F4_4of4-MP2_CRABP_CORE_OF_1TME_VP3_HELIX_TWO.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: MEMYRVR [65 - 71] MP2 NON-HELIX
# VP3 I3-1: LS [72 - 73] INSERTION
# VP3 M-2: DKPHTDD [74 - 80] MP2 HELIX ONE
# VP3 I123-1: INSERTION
# VP3 I3-2: PILCLSLS [81 - 88] INSERTION
# VP3 I23-1: PASD [89 - 92] INSERTION
# VP3 M-3: PR [93 - 94] MP2 HELIX ONE
# VP3 I123-2: INSERTION
# VP3 M-4: LSHT [95 - 98] MP2 HELIX TWO
# VP3 M-5: MLGEILNYYTH [99 - 109] MP2 NON-HELIX
# VP3 I3-3: WA [110 - 111] INSERTION
# VP3 I13-1: GSL [112 - 114] INSERTION
# VP3 B-1: ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: KFTF [115 - 118] ABT CHAIN B
# VP3 I3-4: LFCGSMMATGKLLV [119 - 132] INSERTION
# VP3 B-3: SYA [133 - 135] ABT CHAIN B
# VP3 I1-2: P [136] INSERTION
# VP3 B-4: PGAD [137 - 140] ABT CHAIN B
# VP3 I123-3: P [141] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: PK [142 - 143] INSERTION
# VP3 B-5: KRKEAML [144 - 150] ABT CHAIN B
# VP3 M-6: GTHVIWDIGLQSSCTMVVPWISNTT [151 - 175] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRQTI [176 - 180] INSERTION
# VP3 M-7: DDSFTE [181 - 186] MP2 NON-HELIX
# VP3 I123-5: GGYISVFY [187 - 194] INSERTION
# VP3 M-8: Q [195] MP2 NON-HELIX
# VP3 I123-6: T [196] INSERTION
# VP3 M-9: R [197] MP2 NON-HELIX
# VP3 I23-3: INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: IVV [198 - 200] MP2 NON-HELIX
# VP3 I13-2: PLS [201 - 203] INSERTION
# VP3 I123-7: TPR [204 - 206] INSERTION
# VP3 M-12: EMDILGFVSAC [207 - 217] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: NDFSVRLLR [218 - 226] ABT CHAIN A
# VP3 M-13: DTT [227 - 229] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: HIEQKALA (RES 236-237 'LA' MISSING) [230 - 237] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_E5_F5_1of4-ICOSAHEDRON.pdb;
# VP3 M-1: MEMYRVR [65 - 71] MP2 NON-HELIX
# VP3 M-5: MLGEILNYYTH [99 - 109] MP2 NON-HELIX
# VP3 M-6: GTHVIWDIGLQSSCTMVVPWISNTT [151 - 175] MP2 NON-HELIX
# VP3 M-7: DDSFTE [181 - 186] MP2 NON-HELIX
# VP3 M-8: Q [195] MP2 NON-HELIX
# VP3 M-9: R [197] MP2 NON-HELIX
# VP3 M-10: MP2 NON-HELIX
# VP3 M-11: IVV [198 - 200] MP2 NON-HELIX
# VP3 M-12: EMDILGFVSAC [207 - 217] MP2 NON-HELIX
# VP3 M-13: DTT [227 - 229] MP2 NON-HELIX
# VP3 M-14: HIEQKALA (RES 236-237 'LA' MISSING) [230 - 237] MP2 NON-HELIX
# VP3 M-15: MP2 NON-HELIX
# VP3 M-16: MP2 NON-HELIX
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
C,-64;C,72-98;C,110-150;C,176-180;C,187-194;C,196;C,201-206;C,218-226;A;B;D;\
G,-64;G,72-98;G,110-150;G,176-180;G,187-194;G,196;G,201-206;G,218-226;E;F;H;\
K,-64;K,72-98;K,110-150;K,176-180;K,187-194;K,196;K,201-206;K,218-226;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E5_F5_2of4-MP2_CRABP_CORE_OF_1HXS_VP3_NON_HELIX.pdb;
# VP3 M-2: DKPHTDD [74 - 80] MP2 HELIX ONE
# VP3 M-3: PR [93 - 94] MP2 HELIX ONE
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
C,-73;C,81-92;C,95-;A;B;D;\
G,-73;G,81-92;G,95-;E;F;H;\
K,-73;K,81-92;K,95-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E5_F5_3of4-MP2_CRABP_CORE_OF_1HXS_VP3_HELIX_ONE.pdb;
# VP3 M-4: LSHT [95 - 98] MP2 HELIX TWO
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
C,-94;C,99-;A;B;D;\
G,-94;G,99-;E;F;H;\
K,-94;K,99-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E5_F5_4of4-MP2_CRABP_CORE_OF_1HXS_VP3_HELIX_TWO.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: MERLRVD [64 - 70] MP2 NON-HELIX
# VP3 I3-1: ISVQA [71 - 75] INSERTION
# VP3 M-2: DLD [76 - 78] MP2 HELIX ONE
# VP3 I123-1: Q [79] INSERTION
# VP3 I3-2: LLFNIPLD [80 - 87] INSERTION
# VP3 I23-1: IQLD [88 - 91] INSERTION
# VP3 M-3: GPLRN [92 - 96] MP2 HELIX ONE
# VP3 I123-2: T [97] INSERTION
# VP3 M-4: LVGNISR [98 - 104] MP2 HELIX TWO
# VP3 M-5: YYTH [105 - 108] MP2 NON-HELIX
# VP3 I3-3: WS [109 - 110] INSERTION
# VP3 I13-1: GSLE [111 - 114] INSERTION
# VP3 B-1: ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: MTF [115 - 117] ABT CHAIN B
# VP3 I3-4: MFCGSFMATGKLILC [118 - 132] INSERTION
# VP3 B-3: YT [133 - 134] ABT CHAIN B
# VP3 I1-2: P [135] INSERTION
# VP3 B-4: PGGSC [136 - 140] ABT CHAIN B
# VP3 I123-3: P [141] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: T [142] INSERTION
# VP3 B-5: TRETAML [143 - 149] ABT CHAIN B
# VP3 M-6: GTHIVWDFGLQSSITLIIPWISGSH [150 - 174] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRMFNS [175 - 180] INSERTION
# VP3 M-7: DAKST [181 - 185] MP2 NON-HELIX
# VP3 I123-5: NANVGYVTCFM [186 - 196] INSERTION
# VP3 M-8: Q [197] MP2 NON-HELIX
# VP3 I123-6: T [198] INSERTION
# VP3 M-9: N [199] MP2 NON-HELIX
# VP3 I23-3: INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: LIV [200 - 202] MP2 NON-HELIX
# VP3 I13-2: P [203] INSERTION
# VP3 I123-7: INSERTION
# VP3 M-12: SESSDTCSLIGFIAAK [204 - 219] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: DDFSLRLMR [220 - 228] ABT CHAIN A
# VP3 M-13: DSPD [229 - 232] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: IGQSNHLHGAEAAYQ [233 - 247] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_E6_F6_1of4-ICOSAHEDRON.pdb;
# VP3 M-1: MERLRVD [64 - 70] MP2 NON-HELIX
# VP3 M-5: YYTH [105 - 108] MP2 NON-HELIX
# VP3 M-6: GTHIVWDFGLQSSITLIIPWISGSH [150 - 174] MP2 NON-HELIX
# VP3 M-7: DAKST [181 - 185] MP2 NON-HELIX
# VP3 M-8: Q [197] MP2 NON-HELIX
# VP3 M-9: N [199] MP2 NON-HELIX
# VP3 M-10: MP2 NON-HELIX
# VP3 M-11: LIV [200 - 202] MP2 NON-HELIX
# VP3 M-12: SESSDTCSLIGFIAAK [204 - 219] MP2 NON-HELIX
# VP3 M-13: DSPD [229 - 232] MP2 NON-HELIX
# VP3 M-14: IGQSNHLHGAEAAYQ [233 - 247] MP2 NON-HELIX
# VP3 M-15: MP2 NON-HELIX
# VP3 M-16: MP2 NON-HELIX
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
C,-63;C,71-104;C,109-149;C,175-180;C,186-196;C,198;C,203;C,220-228;A;B;D;\
G,-63;G,71-104;G,109-149;G,175-180;G,186-196;G,198;G,203;G,220-228;E;F;H;\
K,-63;K,71-104;K,109-149;K,175-180;K,186-196;K,198;K,203;K,220-228;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E6_F6_2of4-MP2_CRABP_CORE_OF_4WM7_VP3_NON_HELIX.pdb;
# VP3 M-2: DLD [76 - 78] MP2 HELIX ONE
# VP3 M-3: GPLRN [92 - 96] MP2 HELIX ONE
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
C,-75;C,79-91;C,97-;A;B;D;\
G,-75;G,79-91;G,97-;E;F;H;\
K,-75;K,79-91;K,97-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E6_F6_3of4-MP2_CRABP_CORE_OF_4WM7_VP3_HELIX_ONE.pdb;
# VP3 M-4: LVGNISR [98 - 104] MP2 HELIX TWO
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
C,-97;C,105-;A;B;D;\
G,-97;G,105-;E;F;H;\
K,-97;K,105-;I;J;L;"\
--output=new/figs/fig8/figure_8_panels_E6_F6_4of4-MP2_CRABP_CORE_OF_4WM7_VP3_HELIX_TWO.pdb;
# Combine similar substructures of VP1, VP2, and VP3 to make Figure 8 VP123 substructures.
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_G2_H2_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A2_B2_2of4-MP2_CRABP_CORE_OF_1BBT_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_C2_D2_2of4-MP2_CRABP_CORE_OF_1BBT_VP2_NON_HELIX.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E2_F2_2of4-MP2_CRABP_CORE_OF_1BBT_VP3_NON_HELIX.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G2_H2_2of4-MP2_CRABP_CORE_OF_1BBT_VP123_NON_HELIX.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A2_B2_3of4-MP2_CRABP_CORE_OF_1BBT_VP1_HELIX_ONE.pdb \
--input2=new/figs/fig8/figure_8_panels_C2_D2_3of4-MP2_CRABP_CORE_OF_1BBT_VP2_HELIX_ONE.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E2_F2_3of4-MP2_CRABP_CORE_OF_1BBT_VP3_HELIX_ONE.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G2_H2_3of4-MP2_CRABP_CORE_OF_1BBT_VP123_HELIX_ONE.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A2_B2_4of4-MP2_CRABP_CORE_OF_1BBT_VP1_HELIX_TWO.pdb \
--input2=new/figs/fig8/figure_8_panels_C2_D2_4of4-MP2_CRABP_CORE_OF_1BBT_VP2_HELIX_TWO.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E2_F2_4of4-MP2_CRABP_CORE_OF_1BBT_VP3_HELIX_TWO.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G2_H2_4of4-MP2_CRABP_CORE_OF_1BBT_VP123_HELIX_TWO.pdb;
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_G3_H3_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A3_B3_2of4-MP2_CRABP_CORE_OF_1TMF_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_C3_D3_2of4-MP2_CRABP_CORE_OF_1TMF_VP2_NON_HELIX.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E3_F3_2of4-MP2_CRABP_CORE_OF_1TMF_VP3_NON_HELIX.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G3_H3_2of4-MP2_CRABP_CORE_OF_1TMF_VP123_NON_HELIX.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A3_B3_3of4-MP2_CRABP_CORE_OF_1TMF_VP1_HELIX_ONE.pdb \
--input2=new/figs/fig8/figure_8_panels_C3_D3_3of4-MP2_CRABP_CORE_OF_1TMF_VP2_HELIX_ONE.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E3_F3_3of4-MP2_CRABP_CORE_OF_1TMF_VP3_HELIX_ONE.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G3_H3_3of4-MP2_CRABP_CORE_OF_1TMF_VP123_HELIX_ONE.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A3_B3_4of4-MP2_CRABP_CORE_OF_1TMF_VP1_HELIX_TWO.pdb \
--input2=new/figs/fig8/figure_8_panels_C3_D3_4of4-MP2_CRABP_CORE_OF_1TMF_VP2_HELIX_TWO.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E3_F3_4of4-MP2_CRABP_CORE_OF_1TMF_VP3_HELIX_TWO.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G3_H3_4of4-MP2_CRABP_CORE_OF_1TMF_VP123_HELIX_TWO.pdb;
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_G4_H4_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A4_B4_2of4-MP2_CRABP_CORE_OF_1TME_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_C4_D4_2of4-MP2_CRABP_CORE_OF_1TME_VP2_NON_HELIX.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E4_F4_2of4-MP2_CRABP_CORE_OF_1TME_VP3_NON_HELIX.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G4_H4_2of4-MP2_CRABP_CORE_OF_1TME_VP123_NON_HELIX.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A4_B4_3of4-MP2_CRABP_CORE_OF_1TME_VP1_HELIX_ONE.pdb \
--input2=new/figs/fig8/figure_8_panels_C4_D4_3of4-MP2_CRABP_CORE_OF_1TME_VP2_HELIX_ONE.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E4_F4_3of4-MP2_CRABP_CORE_OF_1TME_VP3_HELIX_ONE.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G4_H4_3of4-MP2_CRABP_CORE_OF_1TME_VP123_HELIX_ONE.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A4_B4_4of4-MP2_CRABP_CORE_OF_1TME_VP1_HELIX_TWO.pdb \
--input2=new/figs/fig8/figure_8_panels_C4_D4_4of4-MP2_CRABP_CORE_OF_1TME_VP2_HELIX_TWO.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E4_F4_4of4-MP2_CRABP_CORE_OF_1TME_VP3_HELIX_TWO.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G4_H4_4of4-MP2_CRABP_CORE_OF_1TME_VP123_HELIX_TWO.pdb;
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_G5_H5_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A5_B5_2of4-MP2_CRABP_CORE_OF_1HXS_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_C5_D5_2of4-MP2_CRABP_CORE_OF_1HXS_VP2_NON_HELIX.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E5_F5_2of4-MP2_CRABP_CORE_OF_1HXS_VP3_NON_HELIX.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G5_H5_2of4-MP2_CRABP_CORE_OF_1HXS_VP123_NON_HELIX.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A5_B5_3of4-MP2_CRABP_CORE_OF_1HXS_VP1_HELIX_ONE.pdb \
--input2=new/figs/fig8/figure_8_panels_C5_D5_3of4-MP2_CRABP_CORE_OF_1HXS_VP2_HELIX_ONE.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E5_F5_3of4-MP2_CRABP_CORE_OF_1HXS_VP3_HELIX_ONE.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G5_H5_3of4-MP2_CRABP_CORE_OF_1HXS_VP123_HELIX_ONE.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A5_B5_4of4-MP2_CRABP_CORE_OF_1HXS_VP1_HELIX_TWO.pdb \
--input2=new/figs/fig8/figure_8_panels_C5_D5_4of4-MP2_CRABP_CORE_OF_1HXS_VP2_HELIX_TWO.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E5_F5_4of4-MP2_CRABP_CORE_OF_1HXS_VP3_HELIX_TWO.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G5_H5_4of4-MP2_CRABP_CORE_OF_1HXS_VP123_HELIX_TWO.pdb;
cp new/tmpicosframe.pdb new/figs/fig8/figure_8_panels_G6_H6_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A6_B6_2of4-MP2_CRABP_CORE_OF_4WM7_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_C6_D6_2of4-MP2_CRABP_CORE_OF_4WM7_VP2_NON_HELIX.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E6_F6_2of4-MP2_CRABP_CORE_OF_4WM7_VP3_NON_HELIX.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G6_H6_2of4-MP2_CRABP_CORE_OF_4WM7_VP123_NON_HELIX.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A6_B6_3of4-MP2_CRABP_CORE_OF_4WM7_VP1_HELIX_ONE.pdb \
--input2=new/figs/fig8/figure_8_panels_C6_D6_3of4-MP2_CRABP_CORE_OF_4WM7_VP2_HELIX_ONE.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E6_F6_3of4-MP2_CRABP_CORE_OF_4WM7_VP3_HELIX_ONE.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G6_H6_3of4-MP2_CRABP_CORE_OF_4WM7_VP123_HELIX_ONE.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A6_B6_4of4-MP2_CRABP_CORE_OF_4WM7_VP1_HELIX_TWO.pdb \
--input2=new/figs/fig8/figure_8_panels_C6_D6_4of4-MP2_CRABP_CORE_OF_4WM7_VP2_HELIX_TWO.pdb \
--nopositionchange --output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_E6_F6_4of4-MP2_CRABP_CORE_OF_4WM7_VP3_HELIX_TWO.pdb \
--nopositionchange \
--output=new/figs/fig8/figure_8_panels_G6_H6_4of4-MP2_CRABP_CORE_OF_4WM7_VP123_HELIX_TWO.pdb;
# Figure 9. Myelin P2 trimers, Alpha-bungarotoxin trimers, Picornavirus Myelin P2/CRABP Cores,
# and Picornavirus Toxin Cores.
#
# (Strip atoms to make Figure 9 substructures of Alpha-bungarotoxin and VP1234 trimers.)
#===================================================================================================
# 2WUT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# M-1: GMSNKFLGTWKLVS [(-1) - 12] MP2 NON-HELIX
# M-2: SENFDDYM [13 - 20] MP2 HELIX ONE
# M-3: KALG [21 - 24] MP2 HELIX ONE
# M-4: VGLATRKLGNLAK [25 - 37] MP2 HELIX TWO
# M-5: PTVIISKKGDIITIRTEST [38 - 56] MP2 NON-HELIX
# M-6: FKNTEISFKLG [57 - 67] MP2 NON-HELIX
# M-7: QEFEETT [68 - 74] MP2 NON-HELIX
# M-8: ADN [75 - 77] MP2 NON-HELIX
# M-9: RKT [78 - 80] MP2 NON-HELIX
# M-10: KS [81 - 82] MP2 NON-HELIX
# M-11: IV [83 - 84] MP2 NON-HELIX
# M-12: TLQRGSLNQVQR [85 - 96] MP2 NON-HELIX
# M-13: WDGKETTI [97 - 104] MP2 NON-HELIX
# M-14: KRKLVNGKMVAECKMK [105 - 120] MP2 NON-HELIX
# M-15: GVVCT [121 - 125] MP2 NON-HELIX
# M-16: RIYEKV [126 - 131] MP2 NON-HELIX
cp new/tmpicosframe.pdb new/figs/fig9/figure_9_panels_A_B_1of5-ICOSAHEDRON.pdb;
cp new/super/2WUT_TRIMER_table5.pdb new/figs/fig9/figure_9_panels_A_B_2of5-MP2_CRABP_CORE_OF_2WUT_MYELIN_TRIMER.pdb;
# Atoms are stripped from Alpha-bungarotoxin trimers to make separate trimer substructures
# of Alpha-bungarotoxin A chains and B chains.
#
# The following is useful for Alpha-bungarotoxin chain identification:
#
# 2ABX trimer chain identifier ordinal: 1 2
# First 2ABX chain identifiers: A B
# Second 2ABX chain identifiers: C D
# Third 2ABX chain identifiers: E F
#===================================================================================================
# 2ABX.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# A A-1: IVCHTTATIPSSAVTCPPGENLCYRKMWCDAFCSSRG [1 - 37] ABT TOXIN CORE
# A A-2: KVVELGCAATCPSKK [38 - 52] ABT TOXIN CORE
# A A-3: PYEEVT [53 - 58] ABT TOXIN CORE
# A A-4: CCSTDKCN [59 - 66] ABT TOXIN CORE
# A A-5: HP [67 - 68] ABT TOXIN CORE
# A A-6: PKRQPG [69 - 74] ABT TOXIN CORE
#
# B B-1: IV [1 - 2] ABT TOXIN CORE
# B B-2: CHTTATIP [3 - 10] ABT TOXIN CORE
# B B-3: SSAVTCP [11 - 17] ABT TOXIN CORE
# B B-4: PGENLCYRKMWC [18 - 29] ABT TOXIN CORE
# B B-5: DAFCSSRGKVVELGCAATC [30 - 48] ABT TOXIN CORE
# B B-X: PSKKPYEEVTCCSTDKCNHPPKRQPG [49 - 74] ABT NON-TOXIC CORE
stripPDB --input=new/super/2ABX_TRIMER_table6.pdb --remove="B;D;F;" \
--output=new/figs/fig9/figure_9_panels_A_B_3of5-2ABX_TRIMER_A_CHAINS_TOXIN_CORE.pdb;
stripPDB --input=new/super/2ABX_TRIMER_table6.pdb --remove="A;B,49-;C;D,49-;E;F,49-;" \
--output=new/figs/fig9/figure_9_panels_A_B_4of5-2ABX_TRIMER_B_CHAINS_TOXIN_CORE.pdb;
stripPDB --input=new/super/2ABX_TRIMER_table6.pdb --remove="A;B,-48;C;D,-48;E;F,-48;" \
--output=new/figs/fig9/figure_9_panels_A_B_5of5-2ABX_TRIMER_B_CHAINS_NON_TOXIN_CORE.pdb;
# Atoms are stripped from VP1234 trimers to make VP1-trimer substructures
# showing regions corresponding to Alpha-bungarotoxin chain A and separately
# corresponding to Alpha-bungarotoxin chain B residues 1 - 48.
#
# The following is useful for VP1234 chain identification (in panels C - L):
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: TTSAGESADPVTTTVENYGG [1 - 20] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: ETQI [21 - 24] MP2 HELIX ONE
# VP1 I123-1: Q [25] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: RR [26 - 27] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHTDVSFIMDR [28 - 38] MP2 HELIX TWO
# VP1 M-5: FVKVTPQNQI [39 - 48] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: N [49] INSERTION
# VP1 B-1: IL [50 - 51] ABT CHAIN B
# VP1 I1-1: INSERTION
# VP1 B-2: ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: ABT CHAIN B
# VP1 I1-2: D [52] INSERTION
# VP1 B-4: LM [53 - 54] ABT CHAIN B
# VP1 I123-3: QVPSHT [55 - 60] INSERTION
# VP1 I1-3: LV [61 - 62] INSERTION
# VP1 I123-4: GG [63 - 64] INSERTION
# VP1 B-5: LLRAS [65 - 69] ABT CHAIN B
# VP1 M-6: TYYFSDLEIAVKH [70 - 82] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: EG [83 - 84] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: D [85] MP2 NON-HELIX
# VP1 I123-6: LT [86 - 87] INSERTION
# VP1 M-9: MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WV [88 - 89] MP2 NON-HELIX
# VP1 I13-2: PNGA [90 - 93] INSERTION
# VP1 I123-7: PE [94 - 95] INSERTION
# VP1 M-12: KALDNTTNPTAYHKAPLTR [96 - 114] MP2 NON-HELIX
# VP1 A-1: LALPYTAPHRVLATVYNGECRYSRNAV (PDB MISSING 'RYSRNAV') [115 - 141] ABT CHAIN A
# VP1 I1-4: PNLRGDLQVLAQ (PDB MISSING 'PNLRGDLQVLAQ') [142 - 153] INSERTION
# VP1 A-2: KVARTLPTSFNY (RES 154 - 156 'KVA' MISSING FROM PDB) [154 - 165] ABT CHAIN A
# VP1 M-13: GAIKAT [166 - 171] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: RVTELLYRMKRAETYCPRPLLA [172 - 193] MP2 NON-HELIX
# VP1 A-3: I [194] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: H [195] ABT CHAIN A
# VP1 M-15: PTEA [196 - 199] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RHKQKIVAPVK (PDB MISSING 'VK') [200 - 210] MP2 NON-HELIX
# VP1 A-6: QTL (PDB MISSING 'QTL') [211 - 213] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig9/figure_9_panels_C_D_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A2_B2_2of4-MP2_CRABP_CORE_OF_1BBT_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_A2_B2_3of4-MP2_CRABP_CORE_OF_1BBT_VP1_HELIX_ONE.pdb \
--nopositionchange \
--output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_A2_B2_4of4-MP2_CRABP_CORE_OF_1BBT_VP1_HELIX_TWO.pdb \
--output=new/figs/fig9/figure_9_panels_C_D_2of4-MP2_CRABP_CORE_OF_1BBT_VP1.pdb;
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,-114;A,142-153;A,166-193;A,196-210;B;C;D;\
E,-114;E,142-153;E,166-193;E,196-210;F;G;H;\
I,-114;I,142-153;I,166-193;I,196-210;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_C_D_3of4-ABT_A_CHAINS_TOXIN_CORE_OF_1BBT_VP1.pdb;
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,-49;A,52;A,55-64;A,70-;B;C;D;\
E,-49;E,52;E,55-64;E,70-;F;G;H;\
I,-49;I,52;I,55-64;I,70-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_C_D_4of4-ABT_B_CHAINS_TOXIN_CORE_OF_1BBT_VP1.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GVDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FNY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDYRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSSSGPQKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGVN [99 - 102] INSERTION
# VP1 I123-4: GAN [103 - 105] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [106 - 120] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 I123-6: ASVL [142 - 145] INSERTION
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [147 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGA [149 - 152] INSERTION
# VP1 I123-7: PAD [153 - 155] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [156 - 176] MP2 NON-HELIX
# VP1 A-1: LVGAGNSQVSFVVPYNSPLSVLPAAWFNGWS [177 - 207] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [209 - 220] INSERTION
# VP1 A-2: ADFGRLWI [221 - 228] ABT CHAIN A
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [251 - 256] MP2 NON-HELIX
# VP1 I2-6: PT [257 - 258] INSERTION
# VP1 M-16: PTTTKINADNPVPILELE [259 - 276] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig9/figure_9_panels_E_F_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A3_B3_2of4-MP2_CRABP_CORE_OF_1TMF_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_A3_B3_3of4-MP2_CRABP_CORE_OF_1TMF_VP1_HELIX_ONE.pdb \
--nopositionchange \
--output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_A3_B3_4of4-MP2_CRABP_CORE_OF_1TMF_VP1_HELIX_TWO.pdb \
--output=new/figs/fig9/figure_9_panels_E_F_2of4-MP2_CRABP_CORE_OF_1TMF_VP1.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-176;A,209-220;A,229-;B;C;D;\
E,-176;E,209-220;E,229-;F;G;H;\
I,-176;I,209-220;I,229-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_E_F_3of4-ABT_A_CHAINS_TOXIN_CORE_OF_1TMF_VP1.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-66;A,74;A,78-89;A,96-105;A,121-;B;C;D;\
E,-66;E,74;E,78-89;E,96-105;E,121-;F;G;H;\
I,-66;I,74;I,78-89;I,96-105;I,121-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_E_F_4of4-ABT_B_CHAINS_TOXIN_CORE_OF_1TMF_VP1.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GSDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNIEST [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FVY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDLRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSTSGPVKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGT [99 - 101] INSERTION
# VP1 I123-4: TN [102 - 103] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [104 - 118] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [119 - 134] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [135 - 138] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [139] MP2 NON-HELIX
# VP1 I123-6: ASVL [140 - 143] INSERTION
# VP1 M-9: R [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [145 - 146] MP2 NON-HELIX
# VP1 I13-2: PTGA [147 - 150] INSERTION
# VP1 I123-7: PAD [151 - 153] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [154 - 174] MP2 NON-HELIX
# VP1 A-1: LVGAGNTQISFVVPYNSPLSVLPAAWFNGWS [175 - 205] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [206 - 218] INSERTION
# VP1 A-2: ADFGRLWI [219 - 226] ABT CHAIN A
# VP1 M-13: QGNTSA [227 - 232] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [233 - 248] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [249 - 254] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: PVSTRSKINADNPVPILELE [255 - 274] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig9/figure_9_panels_G_H_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A4_B4_2of4-MP2_CRABP_CORE_OF_1TME_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_A4_B4_3of4-MP2_CRABP_CORE_OF_1TME_VP1_HELIX_ONE.pdb \
--nopositionchange \
--output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_A4_B4_4of4-MP2_CRABP_CORE_OF_1TME_VP1_HELIX_TWO.pdb \
--output=new/figs/fig9/figure_9_panels_G_H_2of4-MP2_CRABP_CORE_OF_1TME_VP1.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,-174;A,206-218;A,227-;B;C;D;\
E,-174;E,206-218;E,227-;F;G;H;\
I,-174;I,206-218;I,227-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_G_H_3of4-ABT_A_CHAINS_TOXIN_CORE_OF_1TME_VP1.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,-66;A,74;A,78-89;A,96-103;A,119-;B;C;D;\
E,-66;E,74;E,78-89;E,96-103;E,119-;F;G;H;\
I,-66;I,74;I,78-89;I,96-103;I,119-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_G_H_4of4-ABT_B_CHAINS_TOXIN_CORE_OF_1TME_VP1.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PALTAVETGATNPLV [43 - 56] MP2 NON-HELIX
# VP1 I3-1: P [57] INSERTION
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 I123-1: Q [62] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHRSRS [68 - 73] MP2 HELIX TWO
# VP1 M-5: ESSIESFFARGACVTIMT [74 - 91] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: V [92] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: DN [93 - 94] INSERTION
# VP1 B-2: PAS [95 - 97] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: TT [98 - 99] ABT CHAIN B
# VP1 I1-2: NKDK [100 - 103] INSERTION
# VP1 B-4: LFAVW [104 - 108] ABT CHAIN B
# VP1 I123-3: K [109] INSERTION
# VP1 I1-3: IT [110 - 111] INSERTION
# VP1 I123-4: YK [112 - 113] INSERTION
# VP1 B-5: DTVQLRRKLEF [114 - 124] ABT CHAIN B
# VP1 M-6: FTYSRFDMELTFVVTA [125 - 140] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NFTETNN [141 - 147] MP2 NON-HELIX
# VP1 I123-5: GHA [148 - 150] INSERTION
# VP1 M-8: LNQ [151 - 153] MP2 NON-HELIX
# VP1 I123-6: VY [154 - 155] INSERTION
# VP1 M-9: Q [156] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: IMYV [157 - 160] MP2 NON-HELIX
# VP1 I13-2: PPGA [161 - 164] INSERTION
# VP1 I123-7: PVPE [165 - 168] INSERTION
# VP1 M-12: KWDDYTWQTSSNPSIFYTYGTAPAR [169 - 193] MP2 NON-HELIX
# VP1 A-1: ISVPYVGISNAYSHFYDGFSKV [194 - 215] ABT CHAIN A
# VP1 I1-4: PLKDQSAALGDSLYGAASLN [216 - 235] INSERTION
# VP1 A-2: DFGILAVRV [236 - 244] ABT CHAIN A
# VP1 M-13: VNDHNPT [245 - 251] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: KVTSKIRVYLKPKHIRVWCPRPPRAVA [252 - 278] MP2 NON-HELIX
# VP1 A-3: YYG [279 - 281] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: PGVDY [282 - 286] ABT CHAIN A
# VP1 I1-5: KD [287 - 288] INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: GTLTP [289 - 293] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: LSTKD [294 - 298] MP2 NON-HELIX
# VP1 A-6: LTTY [299 - 302] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig9/figure_9_panels_I_J_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A5_B5_2of4-MP2_CRABP_CORE_OF_1HXS_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_A5_B5_3of4-MP2_CRABP_CORE_OF_1HXS_VP1_HELIX_ONE.pdb \
--nopositionchange \
--output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_A5_B5_4of4-MP2_CRABP_CORE_OF_1HXS_VP1_HELIX_TWO.pdb \
--output=new/figs/fig9/figure_9_panels_I_J_2of4-MP2_CRABP_CORE_OF_1HXS_VP1.pdb;
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-193;A,216-235;A,245-278;A,287-298;B;C;D;\
E,-193;E,216-235;E,245-278;E,287-298;F;G;H;\
I,-193;I,216-235;E,245-278;I,287-298;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_I_J_3of4-ABT_A_CHAINS_TOXIN_CORE_OF_1HXS_VP1.pdb;
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-94;A,100-103;A,96-103;A,109-;B;C;D;\
E,-94;E,100-103;E,96-103;E,109-;F;G;H;\
I,-94;I,100-103;I,96-103;I,109-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_I_J_4of4-ABT_B_CHAINS_TOXIN_CORE_OF_1HXS_VP1.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig9/figure_9_panels_K_L_1of4-ICOSAHEDRON.pdb;
combinePDB --input1=new/figs/fig8/figure_8_panels_A6_B6_2of4-MP2_CRABP_CORE_OF_4WM7_VP1_NON_HELIX.pdb \
--input2=new/figs/fig8/figure_8_panels_A6_B6_3of4-MP2_CRABP_CORE_OF_4WM7_VP1_HELIX_ONE.pdb \
--nopositionchange \
--output=new/tmp.pdb;
combinePDB --input1=new/tmp.pdb \
--input2=new/figs/fig8/figure_8_panels_A6_B6_4of4-MP2_CRABP_CORE_OF_4WM7_VP1_HELIX_TWO.pdb \
--output=new/figs/fig9/figure_9_panels_K_L_2of4-MP2_CRABP_CORE_OF_4WM7_VP1.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-182;A,202-214;A,225-257;A,268-271;A,274-290;B;C;D;\
E,-182;E,202-214;E,225-257;E,268-271;E,274-290;F;G;H;\
I,-182;I,202-214;I,225-257;I,268-271;I,274-290;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_K_L_3of4-ABT_A_CHAINS_TOXIN_CORE_OF_4WM7_VP1.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-78;A,85-88;A,94-98;A,110-;B;C;D;\
E,-78;E,85-88;E,94-98;E,110-;F;G;H;\
I,-78;I,85-88;I,94-98;I,110-;J;K;L;"\
--output=new/figs/fig9/figure_9_panels_K_L_4of4-ABT_B_CHAINS_TOXIN_CORE_OF_4WM7_VP1.pdb;
# Figure 10. Toxin Domain Structure in Alpha-bungarotoxin and EV-D68.
#
# Strip atoms to make Figure 10 substructures of prealigned Alpha-bungarotoxin and
# 4WM7 VP1234 trimers.
#
# Atoms are stripped from Alpha-bungarotoxin trimers to show a complimentary-charged toxin annulus
# (i.e., the "TOX DOMAIN").
#
# The following is useful for Alpha-bungarotoxin chain identification:
#
# 2ABX trimer chain identifier ordinal: 1 2
# First 2ABX chain identifiers: A B
# Second 2ABX chain identifiers: C D
# Third 2ABX chain identifiers: E F
#
# Atoms are stripped from 4WM7 (EV-D68) VP1234 trimers to show residues corresponding
# to specific TOX DOMAIN Alpha-bungarotoxin residues.
# The following is useful for VP1234 chain identification (in panels C - L):
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#===================================================================================================
# 2ABX.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# A A-1: IVCHTTATIPSSAVTCPPGENLCYRKMWCDAFCSSRG [1 - 37] ABT TOXIN CORE
# A A-2: KVVELGCAATCPSKK [38 - 52] ABT TOXIN CORE
# A A-3: PYEEVT [53 - 58] ABT TOXIN CORE
# A A-4: CCSTDKCN [59 - 66] ABT TOXIN CORE
# A A-5: HP [67 - 68] ABT TOXIN CORE
# A A-6: PKRQPG [69 - 74] ABT TOXIN CORE
#
# B B-1: IV [1 - 2] ABT TOXIN CORE
# B B-2: CHTTATIP [3 - 10] ABT TOXIN CORE
# B B-3: SSAVTCP [11 - 17] ABT TOXIN CORE
# B B-4: PGENLCYRKMWC [18 - 29] ABT TOXIN CORE
# B B-5: DAFCSSRGKVVELGCAATC [30 - 48] ABT TOXIN CORE
# B B-X: PSKKPYEEVTCCSTDKCNHPPKRQPG [49 - 74] ABT NON-TOXIC CORE
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_A_1of3-ICOSAHEDRON.pdb;
cp new/super/2ABX_TRIMER_table6.pdb new/figs/fig10/figure_10_panel_A_3of3-2ABX_TRIMER.pdb;
# ABT Toxin Loop Residues:
#
# G19-E20-N21-L22-C23 (Figure 1 Section A-1)
# T62-D63-K64-N66 (Figure 1 Section A-4);
# D30-A31-F32 (Figure 1 Section A-1);
# Y54-E55-E56 (Figure 1 Section A-3)
stripPDB --input=new/super/2ABX_TRIMER_table6.pdb --remove="\
A,-18;A,24-29;A,33-53;A,57-61;A,65;A,67-;B;\
C,-18;C,24-29;C,33-53;C,57-61;C,65;C,67-;D;\
E,-18;E,24-29;E,33-53;E,57-61;E,65;E,67-;F;"\
--output=new/figs/fig10/figure_10_panel_A_2of3-2ABX_TRIMER_A_CHAINS_TOXIN_ANNULUS.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_B_1of2-ICOSAHEDRON.pdb;
cp new/figs/fig10/figure_10_panel_A_2of3-2ABX_TRIMER_A_CHAINS_TOXIN_ANNULUS.pdb \
new/figs/fig10/figure_10_panel_B_2of2-2ABX_TRIMER_A_CHAINS_TOXIN_ANNULUS.pdb;
# Strip atoms to make Figure 10 VP12 substructures.
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: S (RES 1 'S' MISSING FROM PDB) [1] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: PSAEA (RES 2-6 'PSAEA' MISSING FROM PDB) [2 - 6] MP2 HELIX ONE
# VP2 I123-1: INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: CGYSD (RES 7-9 'CGY' MISSING FROM PDB) [7 - 11] INSERTION
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LQLKLG [14 - 19] MP2 HELIX TWO
# VP2 M-5: NSAIVTQEAANYC [20 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [33] ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: AY [34 - 35] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GEW [36 - 38] ABT CHAIN B
# VP2 I123-3: P [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: N [40] INSERTION
# VP2 B-5: YLPD [41 - 44] ABT CHAIN B
# VP2 M-6: HEAVAIDK [45 - 52] MP2 NON-HELIX
# VP2 I2-1: PTQPETST [53 - 60] INSERTION
# VP2 I23-2: DRFYTLRSVKW [61 - 71] INSERTION
# VP2 M-7: ESNST [72 - 76] MP2 NON-HELIX
# VP2 I123-5: GWWWKL [77 - 82] INSERTION
# VP2 M-8: PD [83 - 84] MP2 NON-HELIX
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 M-9: NN [87 - 88] MP2 NON-HELIX
# VP2 I23-3: IGMFGQNVQYHYLY [89 - 102] INSERTION
# VP2 I2-2: RS [103 - 104] INSERTION
# VP2 M-10: GFLIHVQCNATKFHQGALL [105 - 123] MP2 NON-HELIX
# VP2 I2-3: INSERTION
# VP2 M-11: VVAI [124 - 127] MP2 NON-HELIX
# VP2 I13-2: P [128] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EHQRGAHDTT [129 - 138] MP2 NON-HELIX
# VP2 A-1: TSPGF [139 - 143] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: NDIMK [144 - 148] ABT CHAIN A
# VP2 M-13: GERGGTF [149 - 155] MP2 NON-HELIX
# VP2 I2-4: N [156] INSERTION
# VP2 M-14: HPYVL [157 - 161] MP2 NON-HELIX
# VP2 A-3: DDGTS [162 - 166] ABT CHAIN A
# VP2 I2-5: IACATIFPHQWINLRTNNSATIVLPWMNVAP [167 - 197] INSERTION
# VP2 A-4: MDFPL [198 - 202] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: RH [203 - 204] ABT CHAIN A
# VP2 M-15: NQWTLAVIPVV [205 - 215] MP2 NON-HELIX
# VP2 I2-6: PLGTRTMSSVVPITVSIAPMCCEFNG [216 - 241] INSERTION
# VP2 M-16: LRHAIT [242 - 247] MP2 NON-HELIX
# VP2 A-6: Q [248] ABT CHAIN A
# Atoms are stripped from 4WM7 (EV-D68) VP1234 trimers to show residues corresponding
# to specific TOX DOMAIN Alpha-bungarotoxin residues.
# The following is useful for VP1234 chain identification (in panels C - L):
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# Panels C, D, E, F, H, I, and J show EV-D68 residues:
#
# VP1 Section I1-2 Subsection 3 residues-HKN ............... H87-K88-N89 (chain A)
# (non-pathogenic sequence in 4WM7.PDB where "TOXLOOP1"
# would be present in pathogenic EV-D68)
# VP1 Section A-1 residues DGF ("TOXLOOP2") ................. D198-G199-F200 (chain A)
# VP1 Section A-2 residues GNLC ("TOXLOOP3") ................ G218-N219-L220-C221 (chain A)
# VP2 Section A-3 Subsection 2 residues ("TOXLOOP4") ........ D162-D163 (chain B)
# VP2 Section A-3 residues .................................. D162-D163-G164-T165-S166 (chain B)
#
# 4WM7.pdb (EV-D68) does not include the chain A residues 81 - 86.
# Panels E - L show the EV-D68 residues (A80,H87-K88-N89) bracketing these missing TOXLOOP residues.
#
# 4WM7.pdb (EV-D68) does not include the chain A residues 212 - 215.
# Panels I and J show the EV-D68 residues (G210-I211,T217-I218) bracketing these missing TOXLOOP residues.
# Panels J,K,L show the EV-D68 residues (I211,T217) immediately bracketing these missing TOXLOOP residues.
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_C_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig4/figure_4_panel_Q3-4WM7_VP1_TRIMER.pdb new/figs/fig10/figure_10_panel_C_3of3-4WM7_VP1_TRIMER.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-86;A,90-197;A,201-217;A,222-;B,-161;B,167-;C;D;\
E,-86;E,90-197;E,201-217;E,222-;F,-161;F,167-;G;H;\
I,-86;I,90-197;I,201-217;I,222-;J,-161;J,167-;K;L;"\
--output=new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_D_1of2-ICOSAHEDRON.pdb;
cp new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb \
new/figs/fig10/figure_10_panel_D_2of2-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_E_1of4-ICOSAHEDRON.pdb;
cp new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb \
new/figs/fig10/figure_10_panel_E_2of4-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/figs/fig4/figure_4_panel_Q3-4WM7_VP1_TRIMER.pdb new/figs/fig10/figure_10_panel_E_3of4-4WM7_VP1_TRIMER.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-79;A,81-86;A,90-;B;C;D;\
E,-79;E,81-86;E,90-;F;G;H;\
I,-79;I,81-86;I,90-;J;K;L;"\
--output=new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_F_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb \
new/figs/fig10/figure_10_panel_F_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb \
new/figs/fig10/figure_10_panel_F_3of3-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_G_1of5-ICOSAHEDRON.pdb;
cp new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb \
new/figs/fig10/figure_10_panel_G_2of5-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/figs/fig4/figure_4_panel_Q3-4WM7_VP1_TRIMER.pdb new/figs/fig10/figure_10_panel_G_3of5-4WM7_VP1_TRIMER.pdb;
cp new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb \
new/figs/fig10/figure_10_panel_G_4of5-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
twwistPDB --exact orig/4WM7.pdb 5123 1079 3154 orig/CASOG_ONE_CAPSID_POINTS.pdb 3 1 2 > new/super/4WM7_tile_1a_with_HETATM.pdb;
twwistPDB --exact orig/4WM7.pdb 5123 1079 3154 orig/CASOG_ONE_CAPSID_POINTS.pdb 6 4 5 > new/super/4WM7_tile_1b_with_HETATM.pdb;
twwistPDB --exact orig/4WM7.pdb 5123 1079 3154 orig/CASOG_ONE_CAPSID_POINTS.pdb 9 7 8 > new/super/4WM7_tile_1c_with_HETATM.pdb;
stripPDB --input=new/super/4WM7_tile_1a_with_HETATM.pdb --output=new/super/PLECONARIL_from_4WM7_1a.pdb --remove="A,-300;A,302-;B;C;D;";
stripPDB --input=new/super/4WM7_tile_1b_with_HETATM.pdb --output=new/super/PLECONARIL_from_4WM7_1b.pdb --remove="A,-300;A,302-;B;C;D;";
stripPDB --input=new/super/4WM7_tile_1c_with_HETATM.pdb --output=new/super/PLECONARIL_from_4WM7_1c.pdb --remove="A,-300;A,302-;B;C;D;";
chainidPDB --input=new/super/PLECONARIL_from_4WM7_1b.pdb --output=new/super/PLECONARIL_from_4WM7_1b_chain_E.pdb --from="A" --to="E";
chainidPDB --input=new/super/PLECONARIL_from_4WM7_1c.pdb --output=new/super/PLECONARIL_from_4WM7_1c_chain_I.pdb --from="A" --to="I";
combinePDB --input1=new/super/PLECONARIL_from_4WM7_1a.pdb --input2=new/super/PLECONARIL_from_4WM7_1b_chain_E.pdb --noidchange \
--output=new/super/PLECONARIL_from_4WM7_1ab.pdb;
combinePDB --input1=new/super/PLECONARIL_from_4WM7_1ab.pdb --input2=new/super/PLECONARIL_from_4WM7_1c_chain_I.pdb --noidchange \
--output=new/figs/fig10/figure_10_panel_G_5of5-4WM7_PLECONARIL_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_H_1of4-ICOSAHEDRON.pdb;
cp new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb \
new/figs/fig10/figure_10_panel_H_2of4-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb \
new/figs/fig10/figure_10_panel_H_3of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
cp new/figs/fig10/figure_10_panel_G_5of5-4WM7_PLECONARIL_TRIMER.pdb \
new/figs/fig10/figure_10_panel_H_4of4-4WM7_PLECONARIL_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panels_I_J_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/figs/fig10/figure_10_panel_C_2of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb \
--remove="A,218;E,218;I,218;" \
--output=new/figs/fig10/figure_10_panels_I_J_2of5-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
cp new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb \
new/figs/fig10/figure_10_panels_I_J_3of5-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-209;A,212-216;A,219-;B;C;D;\
E,-209;E,212-216;E,219-;F;G;H;\
I,-209;I,212-216;I,219-;J;K;L;"\
--output=new/figs/fig10/figure_10_panels_I_J_4of5-RESIDUES_BRACKETING_SECOND_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
cp new/figs/fig10/figure_10_panel_G_5of5-4WM7_PLECONARIL_TRIMER.pdb \
new/figs/fig10/figure_10_panels_I_J_5of5-4WM7_PLECONARIL_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_K_1of5-ICOSAHEDRON.pdb;
cp new/figs/fig4/figure_4_panel_Q3-4WM7_VP1_TRIMER.pdb new/figs/fig10/figure_10_panel_K_2of5-4WM7_TRIMER.pdb;
cp new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb \
new/figs/fig10/figure_10_panel_K_3of5-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-210;A,212-216;A,218-;B;C;D;\
E,-210;E,212-216;E,218-;F;G;H;\
I,-210;I,212-216;I,218-;J;K;L;"\
--output=new/figs/fig10/figure_10_panel_K_4of5-RESIDUES_BRACKETING_SECOND_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb;
cp new/figs/fig10/figure_10_panels_I_J_5of5-4WM7_PLECONARIL_TRIMER.pdb new/figs/fig10/figure_10_panel_K_5of5-4WM7_PLECONARIL_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig10/figure_10_panel_L_1of5-ICOSAHEDRON.pdb;
stripPDB --remove="BCD" --input=new/super/4WM7_tile_1a.pdb --output=new/figs/fig10/figure_10_panel_L_2of5-4WM7_VP1_MONOMER.pdb;
stripPDB --remove="EI" \
--input=new/figs/fig10/figure_10_panel_E_4of4-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb \
--output=new/figs/fig10/figure_10_panel_L_3of5-RESIDUES_BRACKETING_FIRST_MISSING_TOX_DOMAIN_RESIDUES_4WM7_VP1_MONOMER.pdb;
stripPDB --input=new/figs/fig10/figure_10_panels_I_J_4of5-RESIDUES_BRACKETING_SECOND_MISSING_TOX_DOMAIN_RESIDUES_4WM7_TRIMER.pdb --remove="EI" \
--output=new/figs/fig10/figure_10_panel_L_4of5-RESIDUES_BRACKETING_SECOND_MISSING_TOX_DOMAIN_RESIDUES_4WM7_VP1_MONOMER.pdb;
mv new/super/PLECONARIL_from_4WM7_1a.pdb new/figs/fig10/figure_10_panel_L_5of5-4WM7_PLECONARIL_MONOMER.pdb;
rm new/super/4WM7_tile_1a_with_HETATM.pdb;
rm new/super/4WM7_tile_1b_with_HETATM.pdb;
rm new/super/4WM7_tile_1c_with_HETATM.pdb;
rm new/super/PLECONARIL_from_4WM7_1b.pdb;
rm new/super/PLECONARIL_from_4WM7_1c.pdb;
rm new/super/PLECONARIL_from_4WM7_1b_chain_E.pdb;
rm new/super/PLECONARIL_from_4WM7_1c_chain_I.pdb;
rm new/super/PLECONARIL_from_4WM7_1ab.pdb;
# Figure 11. Sialic Acid Binding Domain Components on EV-D68 Capsid.
# (Strip atoms to make Figure 11 substructures.)
# Atoms are stripped from N6 neuraminidase tetramer (1W1X.pdb) to show N6 monomer as cartoon.
stripPDB --input=new/1W1Xx.pdb --remove="BCD" \
--output=new/figs/fig11/figure_11_panel_A_1of2-N6_NEURAMINIDASE_MONOMER.pdb;
# Atoms are stripped from N6 neuraminidase monomer to show the following
# key active site residues of influenza N6 neuraminidase as spheres:
#
# R124, E125, D157, R158, W185, G283, G284, R299, R378, S400, G401, and Y412.
stripPDB --input=new/figs/fig11/figure_11_panel_A_1of2-N6_NEURAMINIDASE_MONOMER.pdb \
--output=new/figs/fig11/figure_11_panel_A_2of2-KEY_N6_SIALIC_ACID_BINDING_RESIDUES.pdb \
--remove="A,-123;A,126-156;A,159-184;A,186-282;A,285-298;A,300-377;A,379-399;A,402-411;A,413-;";
cp new/figs/fig11/figure_11_panel_A_1of2-N6_NEURAMINIDASE_MONOMER.pdb \
new/figs/fig11/figure_11_panel_B_1of3-N6_NEURAMINIDASE_MONOMER.pdb;
cp new/figs/fig11/figure_11_panel_A_2of2-KEY_N6_SIALIC_ACID_BINDING_RESIDUES.pdb \
new/figs/fig11/figure_11_panel_B_2of3-KEY_N6_SIALIC_ACID_BINDING_RESIDUES.pdb;
# Atoms are stripped from the original 1W1X.pdb, which includes HETATMs, to show bound sialic acid.
stripPDB --input=orig/1W1X.pdb --output=new/figs/fig11/figure_11_panel_B_3of3-SIALIC_ACID.pdb \
--remove="A,-1476;A,1478-;B;C;D;" --water --hydrogen;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
# Atoms are stripped from 4WM7 (EV-D68) VP1234 trimers to show residues corresponding
# to specific "SIA BINDING SITE COMPONENT POSITION (WEININGER)".
# The following is useful for VP1234 chain identification (in panels C - L):
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# Panel C shows "SIA BINDING SITE COMPONENT POSITION (WEININGER)" residues:
#
# VP1 Section I13-1 residues-EY ............................. E75-Y76 (chain A)
# VP1 Section I1-1 residue-(E/D) ........................... (chain A)
# VP1 Section M-8 residue-D ............................... D140 (chain A)
# VP1 Section M-12 residue-W ............................... W163 (chain A)
# VP1 Section A-2 residue-R ............................... R223 (chain A)
# VP1 Section M-13 residue-E ............................... E227 (chain A)
# VP1 Section I1-5 residue-R ............................... R270 (chain A)
# VP1 Section M-16 residue-R ............................... R284 (chain A)
stripPDB --input=new/super/4WM7_tile_1a.pdb --remove="BCD" \
--output=new/figs/fig11/figure_11_panel_C_1of2-EVD68_VP1_MONOMER.pdb;
stripPDB --input=new/figs/fig11/figure_11_panel_C_1of2-EVD68_VP1_MONOMER.pdb \
--output=new/figs/fig11/figure_11_panel_C_2of2-EVD68_VP1_MONOMER_PUTATIVE_SIALIC_ACID_BINDING_RESIDUES.pdb \
--remove="A,-74;A,77-139;A,141-162;A,164-222;A,224-226;A,228-269;A,271-283;A,285-;B;C;D;";
cp new/figs/fig11/figure_11_panel_C_1of2-EVD68_VP1_MONOMER.pdb \
new/figs/fig11/figure_11_panel_D_1of3-EVD68_VP1_MONOMER.pdb;
cp new/figs/fig11/figure_11_panel_C_2of2-EVD68_VP1_MONOMER_PUTATIVE_SIALIC_ACID_BINDING_RESIDUES.pdb \
new/figs/fig11/figure_11_panel_D_2of3-EVD68_VP1_MONOMER_PUTATIVE_SIALIC_ACID_BINDING_RESIDUES.pdb;
# HETATM 1 O1A MDL E 1 3.370 83.618 97.618 1.00 0.00 O
# HETATM 2 C9 MDL E 1 5.888 84.723 93.426 1.00 0.00 C
# HETATM 3 C11 MDL E 1 11.270 80.946 95.826 1.00 0.00 C
# Model positioning of figure_11_panel_D_3of3-MODEL_SIALIC_ACID.pdb;
#
# Create PDB file to contain 3 new model atom positions for MODEL_SIALIC_ACID.
printf 'REMARK 250 THREE MODEL ATOMS FOR POSITIONING SIALIC ACID IN FIGURE 11 PANEL D. \n' > new/tmpm;
printf 'HETATM 1 O1A MDL E 1 3.370 83.618 97.618 1.00 0.00 O \n' >> new/tmpm;
printf 'HETATM 2 C9 MDL E 1 5.888 84.723 93.426 1.00 0.00 C \n' >> new/tmpm;
printf 'HETATM 3 C11 MDL E 1 11.270 80.946 95.826 1.00 0.00 C \n' >> new/tmpm;
printf 'END \n' >> new/tmpm;
mv new/tmpm new/figs/fig11//model_sialic_acid_atom_positions.pdb;
# Create figure_11_panel_D_3of3-MODEL_SIALIC_ACID.pdb by positioning
# figure_11_panel_B_3of3-SIALIC_ACID.pdb.
# Three atoms of figure_11_panel_B_3of3-SIALIC_ACID.pdb are superposed with
# model atoms in model_sialic_acid_atom_positions.pdb.
twwistPDB --output=new/figs/fig11/figure_11_panel_D_3of3-MODEL_SIALIC_ACID.pdb \
new/figs/fig11/figure_11_panel_B_3of3-SIALIC_ACID.pdb 12050 12046 12048 \
new/figs/fig11//model_sialic_acid_atom_positions.pdb 1 2 3;
stripPDB --input=new/super/4WM7_tile_1.pdb \
--output=new/figs/fig11/figure_11_panel_E_1of2-EVD68_VP1_TRIMER.pdb \
--remove="B;C;D;F;G;H;J;K;L;";
stripPDB --input=new/figs/fig11/figure_11_panel_E_1of2-EVD68_VP1_TRIMER.pdb \
--output=new/figs/fig11/figure_11_panel_E_2of2-EVD68_VP1_TRIMER_PUTATIVE_SIALIC_ACID_BINDING_RESIDUES.pdb \
--remove="\
A,-74;A,77-139;A,141-162;A,164-222;A,224-226;A,228-269;A,271-283;A,285-;B;C;D;\
E,-74;E,77-139;E,141-162;E,164-222;E,224-226;E,228-269;E,271-283;E,285-;F;G;H;\
I,-74;I,77-139;I,141-162;I,164-222;I,224-226;I,228-269;I,271-283;I,285-;J;K;L;";
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: S (RES 1 'S' MISSING FROM PDB) [1] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: PSAEA (RES 2-6 'PSAEA' MISSING FROM PDB) [2 - 6] MP2 HELIX ONE
# VP2 I123-1: INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: CGYSD (RES 7-9 'CGY' MISSING FROM PDB) [7 - 11] INSERTION
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LQLKLG [14 - 19] MP2 HELIX TWO
# VP2 M-5: NSAIVTQEAANYC [20 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [33] ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: AY [34 - 35] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GEW [36 - 38] ABT CHAIN B
# VP2 I123-3: P [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: N [40] INSERTION
# VP2 B-5: YLPD [41 - 44] ABT CHAIN B
# VP2 M-6: HEAVAIDK [45 - 52] MP2 NON-HELIX
# VP2 I2-1: PTQPETST [53 - 60] INSERTION
# VP2 I23-2: DRFYTLRSVKW [61 - 71] INSERTION
# VP2 M-7: ESNST [72 - 76] MP2 NON-HELIX
# VP2 I123-5: GWWWKL [77 - 82] INSERTION
# VP2 M-8: PD [83 - 84] MP2 NON-HELIX
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 M-9: NN [87 - 88] MP2 NON-HELIX
# VP2 I23-3: IGMFGQNVQYHYLY [89 - 102] INSERTION
# VP2 I2-2: RS [103 - 104] INSERTION
# VP2 M-10: GFLIHVQCNATKFHQGALL [105 - 123] MP2 NON-HELIX
# VP2 I2-3: INSERTION
# VP2 M-11: VVAI [124 - 127] MP2 NON-HELIX
# VP2 I13-2: P [128] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EHQRGAHDTT [129 - 138] MP2 NON-HELIX
# VP2 A-1: TSPGF [139 - 143] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: NDIMK [144 - 148] ABT CHAIN A
# VP2 M-13: GERGGTF [149 - 155] MP2 NON-HELIX
# VP2 I2-4: N [156] INSERTION
# VP2 M-14: HPYVL [157 - 161] MP2 NON-HELIX
# VP2 A-3: DDGTS [162 - 166] ABT CHAIN A
# VP2 I2-5: IACATIFPHQWINLRTNNSATIVLPWMNVAP [167 - 197] INSERTION
# VP2 A-4: MDFPL [198 - 202] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: RH [203 - 204] ABT CHAIN A
# VP2 M-15: NQWTLAVIPVV [205 - 215] MP2 NON-HELIX
# VP2 I2-6: PLGTRTMSSVVPITVSIAPMCCEFNG [216 - 241] INSERTION
# VP2 M-16: LRHAIT [242 - 247] MP2 NON-HELIX
# VP2 A-6: Q [248] ABT CHAIN A
# Atoms are stripped from 4WM7 (EV-D68) VP1234 trimers to show residues corresponding
# to specific "EV-D68 (PATHOGENIC) CAPSID TOXIN-LIKE LOOP RESIDUES".
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
#
# Panel F shows "EV-D68 (PATHOGENIC) CAPSID TOXIN-LIKE LOOP RESIDUES" residues:
#
# VP1 Section I1-1 residues-(D/N) ........................... N78 (chain A)
# (A/T)DKN
# VP1 Section I1-2 Subsection 4 residues-(A/T) K .......... H87-K88 (chain A)
# T86 is MISSING from 4WM7.pdb ^
# VP1 Section B-4 Subsection 1 residues- N .......... N89 (chain A)
#
# VP1 Section A-1 residues DGF .............................. D198-G199-F200 (chain A)
#
# VP1 Section I1-4 residues EKN ............................. E204-K205-N206 (chain A)
#
# VP1 Section A-2 residues GNLC ............................. G218-N219-L220-C221 (chain A)
#
# VP2 Section A-3 Subsection 2 residues-DD .................. D162-D163 (chain B)
#
# EV-D68 (PATHOGENIC) CAPSID TOXIN-LIKE LOOP RESIDUES shown as magenta spheres in Figure 11 Panel F:
#
# chain A: H87-K88-N89, D198-G199-F200, G218-N219-L220-C221
# chain B: D162-D163
cp new/figs/fig11/figure_11_panel_E_1of2-EVD68_VP1_TRIMER.pdb \
new/figs/fig11/figure_11_panel_F_1of3-EVD68_VP1_TRIMER.pdb;
cp new/figs/fig11/figure_11_panel_E_2of2-EVD68_VP1_TRIMER_PUTATIVE_SIALIC_ACID_BINDING_RESIDUES.pdb \
new/figs/fig11/figure_11_panel_F_2of3-EVD68_VP1_TRIMER_PUTATIVE_SIALIC_ACID_BINDING_RESIDUES.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-86;A,90-197;A,201-203;A,207-217;A,222-;B,-161;B,164-;C;D;\
E,-86;E,90-197;E,201-203;E,207-217;E,222-;F,-161;F,164-;G;H;\
I,-86;I,90-197;I,201-203;I,207-217;I,222-;J,-161;J,164-;K;L;"\
--output=new/figs/fig11/figure_11_panel_F_3of3-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-77;A,79-85;A,90-197;A,201-203;A,207-217;A,222-;B,-161;B,164-;C;D;\
E,-77;E,79-85;E,90-197;E,201-203;E,207-217;E,222-;F,-161;F,164-;G;H;\
I,-77;I,79-85;I,90-197;I,201-203;I,207-217;I,222-;J,-161;J,164-;K;L;"\
--output=new/figs/fig11/figure_11_panel_F_related-ABT_TOX_DOMAIN_OF_4WM7_VP1_AND_VP2.pdb;
# Figure 12. Differences in the spatial position of markers in
# constructed TMEV capsid tiling pieces and capsids.
#
# Atoms are stripped from 1TME.pdb (TMEV-1) amd 1TMF (TMEV-2) VP1234 trimers to show
# individual VP protein trimers and residues corresponding to specific markers.
#
# The following is useful for VP1234 chain identification (in panels C - L):
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
# Panel A shows TMEV-1 (1TME.pdb) VP1 protein with:
# VP1 residue markers Y161-T162 (VP1 Section M-12 Subsections 3-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_A_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_A_2of3-1TME_VP1_TRIMER.pdb \
--remove="BCDFGHJKL";
stripPDB --input=new/figs/fig12/figure_12_panel_A_2of3-1TME_VP1_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_A_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
--remove=",-160;,163-;";
# Panel B shows TMEV-2 (1TMF.pdb) VP1 protein with:
# VP1 residue markers Y163-T164 (VP1 Section M-12 Subsections 3-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_B_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_B_2of3-1TMF_VP1_TRIMER.pdb \
--remove="BCDFGHJKL";
stripPDB --input=new/figs/fig12/figure_12_panel_B_2of3-1TMF_VP1_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_B_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
--remove=",-162;,165-;";
# Panel C shows the superposition of Panels A and B.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_C_1of5-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_A_2of3-1TME_VP1_TRIMER.pdb \
new/figs/fig12/figure_12_panel_C_2of5-1TME_VP1_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_B_2of3-1TMF_VP1_TRIMER.pdb \
new/figs/fig12/figure_12_panel_C_3of5-1TMF_VP1_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_A_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_C_4of5-1TME_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_B_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_C_5of5-1TMF_VP1_TRIMER_MARKERS.pdb;
# Panel D shows TMEV-1 (1TME.pdb) VP2 protein with:
# VP2 residue markers R260-H261-E262 (VP2 Section M-16 Subsections 1-M and 3-M)
# VP2 residue markers Y235-A236-S237 (VP2 Section M-15 Subsections 3-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_D_1of4-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_D_2of4-1TME_VP2_TRIMER.pdb \
--remove="ACDEGHIKL";
stripPDB --input=new/figs/fig12/figure_12_panel_D_2of4-1TME_VP2_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_D_3of4-1TME_VP2_TRIMER_MARKERS.pdb \
--remove=",-234;,238-;";
stripPDB --input=new/figs/fig12/figure_12_panel_D_2of4-1TME_VP2_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_D_4of4-1TME_VP2_TRIMER_MARKERS.pdb \
--remove=",-259;,263-;";
# Panel E shows TMEV-2 (1TMF.pdb) VP2 protein with:
# VP2 residue markers R260-H261-E262 (VP2 Section M-16 Subsections 1-M and 3-M)
# VP2 residue markers Y235-A236-T237 (VP2 Section M-15 Subsections 3-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_E_1of4-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_E_2of4-1TMF_VP2_TRIMER.pdb \
--remove="ACDEGHIKL";
stripPDB --input=new/figs/fig12/figure_12_panel_E_2of4-1TMF_VP2_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_E_3of4-1TMF_VP2_TRIMER_MARKERS.pdb \
--remove=",-234;,238-;";
stripPDB --input=new/figs/fig12/figure_12_panel_E_2of4-1TMF_VP2_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_E_4of4-1TMF_VP2_TRIMER_MARKERS.pdb \
--remove=",-259;,263-;";
# Panel F shows the superposition of Panels D and E.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_F_1of7-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_D_2of4-1TME_VP2_TRIMER.pdb \
new/figs/fig12/figure_12_panel_F_2of7-1TME_VP2_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_E_2of4-1TMF_VP2_TRIMER.pdb \
new/figs/fig12/figure_12_panel_F_3of7-1TMF_VP2_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_D_3of4-1TME_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_F_4of7-1TME_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_D_4of4-1TME_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_F_5of7-1TME_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_E_3of4-1TMF_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_F_6of7-1TMF_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_E_4of4-1TMF_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_F_7of7-1TMF_VP2_TRIMER_MARKERS.pdb;
# Panel G shows TMEV-1 (1TME.pdb) VP3 protein with:
# VP3 residue markers N58-S59-N60-N61-K62-R63-Y64
# (VP3 Section M-15 Subsections 2-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_G_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_G_2of3-1TME_VP3_TRIMER.pdb \
--remove="ABDEFHIJL";
stripPDB --input=new/figs/fig12/figure_12_panel_G_2of3-1TME_VP3_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_G_3of3-1TME_VP3_TRIMER_MARKERS.pdb \
--remove=",-57;,65-;";
# Panel H shows TMEV-2 (1TMF.pdb) VP3 protein with:
# VP3 residue markers N58-T59-N60-N61-K62-R63-Y64
# (VP3 Section M-15 Subsections 2-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_H_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_H_2of3-1TMF_VP3_TRIMER.pdb \
--remove="ABDEFHIJL";
stripPDB --input=new/figs/fig12/figure_12_panel_H_2of3-1TMF_VP3_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_H_3of3-1TMF_VP3_TRIMER_MARKERS.pdb \
--remove=",-57;,65-;";
# Panel I shows the superposition of Panels G and H.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_I_1of5-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_G_2of3-1TME_VP3_TRIMER.pdb \
new/figs/fig12/figure_12_panel_I_2of5-1TME_VP3_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_H_2of3-1TMF_VP3_TRIMER.pdb \
new/figs/fig12/figure_12_panel_I_3of5-1TMF_VP3_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_G_3of3-1TME_VP3_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_I_4of5-1TME_VP3_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_H_3of3-1TMF_VP3_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_I_5of5-1TMF_VP3_TRIMER_MARKERS.pdb;
# Panel J shows TMEV-1 (1TME.pdb) VP4 protein with:
# VP1 residue markers Y161-T162 (VP1 Section M-12 Subsections 3-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_J_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_J_2of3-1TME_VP4_TRIMER.pdb \
--remove="ABCEFGIJK";
stripPDB --input=new/figs/fig12/figure_12_panel_A_2of3-1TME_VP1_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_J_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
--remove=",-10;,15-;";
# Panel K shows TMEV-2 (1TMF.pdb) VP4 protein with:
# VP1 residue markers Y163-T164 (VP1 Section M-12 Subsections 3-M and 4-M)
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_K_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig12/figure_12_panel_K_2of3-1TMF_VP4_TRIMER.pdb \
--remove="ABCEFGIJK";
stripPDB --input=new/figs/fig12/figure_12_panel_B_2of3-1TMF_VP1_TRIMER.pdb \
--output=new/figs/fig12/figure_12_panel_K_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
--remove=",-10;,15-;";
# Panel L shows the superposition of Panels J and K.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_L_1of5-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_J_2of3-1TME_VP4_TRIMER.pdb \
new/figs/fig12/figure_12_panel_L_2of5-1TME_VP4_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_K_2of3-1TMF_VP4_TRIMER.pdb \
new/figs/fig12/figure_12_panel_L_3of5-1TMF_VP4_TRIMER.pdb;
cp new/figs/fig12/figure_12_panel_J_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_L_4of5-1TME_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_K_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_L_5of5-1TMF_VP1_TRIMER_MARKERS.pdb;
# Panel M shows all of the TMEV-1 (1TME.pdb) marker spheres.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_M_1of6-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_A_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_M_2of6-1TME_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_D_3of4-1TME_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_M_3of6-1TME_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_D_4of4-1TME_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_M_4of6-1TME_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_G_3of3-1TME_VP3_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_M_5of6-1TME_VP3_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_J_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_M_6of6-1TME_VP1_TRIMER_MARKERS.pdb;
# Panel N shows all of the TMEV-2 (1TMF.pdb) marker spheres.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_N_1of6-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_B_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_N_2of6-1TMF_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_E_3of4-1TMF_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_N_3of6-1TMF_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_E_4of4-1TMF_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_N_4of6-1TMF_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_H_3of3-1TMF_VP3_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_N_5of6-1TMF_VP3_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_K_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_N_6of6-1TMF_VP1_TRIMER_MARKERS.pdb;
# Panel O shows the superposition of Panels M and N.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_O_1of11-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_A_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_2of11-1TME_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_D_3of4-1TME_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_3of11-1TME_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_D_4of4-1TME_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_4of11-1TME_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_G_3of3-1TME_VP3_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_5of11-1TME_VP3_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_J_3of3-1TME_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_6of11-1TME_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_B_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_7of11-1TMF_VP1_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_E_3of4-1TMF_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_8of11-1TMF_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_E_4of4-1TMF_VP2_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_9of11-1TMF_VP2_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_H_3of3-1TMF_VP3_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_10of11-1TMF_VP3_TRIMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_K_3of3-1TMF_VP1_TRIMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_O_11of11-1TMF_VP1_TRIMER_MARKERS.pdb;
# Panel P shows TMEV-1 (1TME.pdb) 3xVP1234 pentamer VP3 structure cartoons and the TMEV-1 marker spheres.
#
# Pentamer chain identifiers as set up in Figure 6, above:
#
# Chain identifier: ABCDEFGHIJKLMNOPQRSTUVWXYZ123456abcdefghijklmnopqrstuvwxyz78
# VP protein: 123412341234123412341234123412341234123412341234123412341234
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_P_1of7-ICOSAHEDRON.pdb;
stripPDB --input=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_P_2of7-1TME_VP3_PENTAMER.pdb \
--remove="ABDEFHIJLMNPQRTUVXYZ2346abdefhijlmnpqrtuvxyz8";
stripPDB --input=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_P_3of7-1TME_VP1_PENTAMER_MARKERS.pdb \
--remove="\
A,-160;A,163-;B;C;D;\
E,-160;E,163-;F;G;H;\
I,-160;I,163-;J;K;L;\
M,-160;M,163-;N;O;P;\
Q,-160;Q,163-;R;S;T;\
U,-160;U,163-;V;W;X;\
Y,-160;Y,163-;Z;1;2;\
3,-160;3,163-;4;5;6;\
a,-160;a,163-;b;c;d;\
e,-160;e,163-;f;g;h;\
i,-160;i,163-;j;k;l;\
m,-160;m,163-;n;o;p;\
q,-160;q,163-;r;s;t;\
u,-160;u,163-;v;w;x;\
y,-160;y,163-;z;7;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_P_4of7-1TME_VP2_PENTAMER_MARKERS.pdb \
--remove="\
B,-234;B,238-;A;C;D;\
F,-234;F,238-;E;G;H;\
J,-234;J,238-;I;K;L;\
N,-234;N,238-;M;O;P;\
R,-234;R,238-;Q;S;T;\
V,-234;V,238-;U;W;X;\
Z,-234;Z,238-;Y;1;2;\
4,-234;4,238-;3;5;6;\
b,-234;b,238-;a;c;d;\
f,-234;f,238-;e;g;h;\
j,-234;j,238-;i;k;l;\
n,-234;n,238-;m;o;p;\
r,-234;r,238-;q;s;t;\
v,-234;v,238-;u;w;x;\
z,-234;z,238-;y;7;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_P_5of7-1TME_VP2_PENTAMER_MARKERS.pdb \
--remove="\
B,-259;B,263-;A;C;D;\
F,-259;F,263-;E;G;H;\
J,-259;J,263-;I;K;L;\
N,-259;N,263-;M;O;P;\
R,-259;R,263-;Q;S;T;\
V,-259;V,263-;U;W;X;\
Z,-259;Z,263-;Y;1;2;\
4,-259;4,263-;3;5;6;\
b,-259;b,263-;a;c;d;\
f,-259;f,263-;e;g;h;\
j,-259;j,263-;i;k;l;\
n,-259;n,263-;m;o;p;\
r,-259;r,263-;q;s;t;\
v,-259;v,263-;u;w;x;\
z,-259;z,263-;y;7;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_P_6of7-1TME_VP3_PENTAMER_MARKERS.pdb \
--remove="\
C,-57;C,65-;A;B;D;\
G,-57;G,65-;E;F;H;\
K,-57;K,65-;I;J;L;\
O,-57;O,65-;M;N;P;\
S,-57;S,65-;Q;R;T;\
W,-57;W,65-;U;V;X;\
1,-57;1,65-;Y;Z;2;\
5,-57;5,65-;3;4;6;\
c,-57;c,65-;a;b;d;\
g,-57;g,65-;e;f;h;\
k,-57;k,65-;i;j;l;\
o,-57;o,65-;m;n;p;\
s,-57;s,65-;q;r;t;\
w,-57;w,65-;u;v;x;\
7,-57;7,65-;y;z;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_A_D_G_J_2of2-1TME_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_P_7of7-1TME_VP1_PENTAMER_MARKERS.pdb \
--remove="\
A,-10;A,15-;B;C;D;\
E,-10;E,15-;F;G;H;\
I,-10;I,15-;J;K;L;\
M,-10;M,15-;N;O;P;\
Q,-10;Q,15-;R;S;T;\
U,-10;U,15-;V;W;X;\
Y,-10;Y,15-;Z;1;2;\
3,-10;3,15-;4;5;6;\
a,-10;a,15-;b;c;d;\
e,-10;e,15-;f;g;h;\
i,-10;i,15-;j;k;l;\
m,-10;m,15-;n;o;p;\
q,-10;q,15-;r;s;t;\
u,-10;u,15-;v;w;x;\
y,-10;y,15-;z;7;8;";
# Panel Q shows TMEV-2 (1TMF.pdb) 3xVP1234 pentamer VP3 structure cartoons and the TMEV-2 marker spheres.
#
# Pentamer chain identifiers as set up in Figure 6, above:
#
# Chain identifier: ABCDEFGHIJKLMNOPQRSTUVWXYZ123456abcdefghijklmnopqrstuvwxyz78
# VP protein: 123412341234123412341234123412341234123412341234123412341234
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_Q_1of7-ICOSAHEDRON.pdb;
stripPDB --input=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_Q_2of7-1TMF_VP3_PENTAMER.pdb \
--remove="ABDEFHIJLMNPQRTUVXYZ2346abdefhijlmnpqrtuvxyz8";
stripPDB --input=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_Q_3of7-1TMF_VP1_PENTAMER_MARKERS.pdb \
--remove="\
A,-162;A,165-;B;C;D;\
E,-162;E,165-;F;G;H;\
I,-162;I,165-;J;K;L;\
M,-162;M,165-;N;O;P;\
Q,-162;Q,165-;R;S;T;\
U,-162;U,165-;V;W;X;\
Y,-162;Y,165-;Z;1;2;\
3,-162;3,165-;4;5;6;\
a,-162;a,165-;b;c;d;\
e,-162;e,165-;f;g;h;\
i,-162;i,165-;j;k;l;\
m,-162;m,165-;n;o;p;\
q,-162;q,165-;r;s;t;\
u,-162;u,165-;v;w;x;\
y,-162;y,165-;z;7;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_Q_4of7-1TMF_VP2_PENTAMER_MARKERS.pdb \
--remove="\
B,-234;B,238-;A;C;D;\
F,-234;F,238-;E;G;H;\
J,-234;J,238-;I;K;L;\
N,-234;N,238-;M;O;P;\
R,-234;R,238-;Q;S;T;\
V,-234;V,238-;U;W;X;\
Z,-234;Z,238-;Y;1;2;\
4,-234;4,238-;3;5;6;\
b,-234;b,238-;a;c;d;\
f,-234;f,238-;e;g;h;\
j,-234;j,238-;i;k;l;\
n,-234;n,238-;m;o;p;\
r,-234;r,238-;q;s;t;\
v,-234;v,238-;u;w;x;\
z,-234;z,238-;y;7;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_Q_5of7-1TMF_VP2_PENTAMER_MARKERS.pdb \
--remove="\
B,-259;B,263-;A;C;D;\
F,-259;F,263-;E;G;H;\
J,-259;J,263-;I;K;L;\
N,-259;N,263-;M;O;P;\
R,-259;R,263-;Q;S;T;\
V,-259;V,263-;U;W;X;\
Z,-259;Z,263-;Y;1;2;\
4,-259;4,263-;3;5;6;\
b,-259;b,263-;a;c;d;\
f,-259;f,263-;e;g;h;\
j,-259;j,263-;i;k;l;\
n,-259;n,263-;m;o;p;\
r,-259;r,263-;q;s;t;\
v,-259;v,263-;u;w;x;\
z,-259;z,263-;y;7;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_Q_6of7-1TMF_VP3_PENTAMER_MARKERS.pdb \
--remove="\
C,-57;C,65-;A;B;D;\
G,-57;G,65-;E;F;H;\
K,-57;K,65-;I;J;L;\
O,-57;O,65-;M;N;P;\
S,-57;S,65-;Q;R;T;\
W,-57;W,65-;U;V;X;\
1,-57;1,65-;Y;Z;2;\
5,-57;5,65-;3;4;6;\
c,-57;c,65-;a;b;d;\
g,-57;g,65-;e;f;h;\
k,-57;k,65-;i;j;l;\
o,-57;o,65-;m;n;p;\
s,-57;s,65-;q;r;t;\
w,-57;w,65-;u;v;x;\
7,-57;7,65-;y;z;8;";
stripPDB --input=new/figs/fig6/figure_6_panels_B_E_H_K_2of2-1TMF_3xVP1234_PENTAMER.pdb \
--output=new/figs/fig12/figure_12_panel_Q_7of7-1TMF_VP1_PENTAMER_MARKERS.pdb \
--remove="\
A,-10;A,15-;B;C;D;\
E,-10;E,15-;F;G;H;\
I,-10;I,15-;J;K;L;\
M,-10;M,15-;N;O;P;\
Q,-10;Q,15-;R;S;T;\
U,-10;U,15-;V;W;X;\
Y,-10;Y,15-;Z;1;2;\
3,-10;3,15-;4;5;6;\
a,-10;a,15-;b;c;d;\
e,-10;e,15-;f;g;h;\
i,-10;i,15-;j;k;l;\
m,-10;m,15-;n;o;p;\
q,-10;q,15-;r;s;t;\
u,-10;u,15-;v;w;x;\
y,-10;y,15-;z;7;8;";
# Panel R shows the superposition between Panels P and Q.
cp new/tmpicosframe.pdb new/figs/fig12/figure_12_panel_R_1of13-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_P_2of7-1TME_VP3_PENTAMER.pdb \
new/figs/fig12/figure_12_panel_R_2of13-1TME_VP3_PENTAMER.pdb;
cp new/figs/fig12/figure_12_panel_P_3of7-1TME_VP1_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_3of13-1TME_VP1_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_P_4of7-1TME_VP2_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_4of13-1TME_VP2_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_P_5of7-1TME_VP2_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_5of13-1TME_VP2_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_P_6of7-1TME_VP3_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_6of13-1TME_VP3_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_P_7of7-1TME_VP1_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_7of13-1TME_VP1_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_Q_2of7-1TMF_VP3_PENTAMER.pdb \
new/figs/fig12/figure_12_panel_R_8of13-1TMF_VP3_PENTAMER.pdb;
cp new/figs/fig12/figure_12_panel_Q_3of7-1TMF_VP1_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_9of13-1TMF_VP1_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_Q_4of7-1TMF_VP2_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_10of13-1TMF_VP2_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_Q_5of7-1TMF_VP2_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_11of13-1TMF_VP2_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_Q_6of7-1TMF_VP3_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_12of13-1TMF_VP3_PENTAMER_MARKERS.pdb;
cp new/figs/fig12/figure_12_panel_Q_7of7-1TMF_VP1_PENTAMER_MARKERS.pdb \
new/figs/fig12/figure_12_panel_R_13of13-1TMF_VP1_PENTAMER_MARKERS.pdb;
# Figure 13. VP3 Protein Beta Sheets circling the pentamer interface
# form the base of a diaphragm shutter-like pore.
#
# Atoms are stripped from 1TME.pdb (TMEV-1) amd 1TMF (TMEV-2) VP1234 trimers to show
# VP3 trimers and proline residue main chain oxygen atoms.
cp new/tmpicosframe.pdb new/figs/fig13/figure_13_panel_A_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_G_2of3-1TME_VP3_TRIMER.pdb new/figs/fig13/figure_13_panel_A_2of3-1TME_VP3_TRIMER.pdb;
stripPDB --input=new/figs/fig12/figure_12_panel_G_2of3-1TME_VP3_TRIMER.pdb \
--output=new/figs/fig13/figure_13_panel_A_3of3-1TME_VP3_TRIMER_PROLINES.pdb \
--remove="\
,,-3998;,,4000-4132;,,4134-4168;,,4170-4236;,,4238-4379;,,4381-4424;,,4426-4493;\
,,4495-4562;,,4564-4996;,,4998-5003;,,5005-5032;,,5034-5243;,,5245-5275;,,5277-5477;\
,,5479-5511;,,5513-5535;,,5537-5685;,,5687-5706;,,5708-5750;,,5752-9954;,,9956-10088;\
,,10090-10124;,,10126-10192;,,10194-10335;,,10337-10380;,,10382-10449;,,10451-10518;\
,,10520-10952;,,10954-10959;,,10961-10988;,,10990-11199;,,11201-11231;,,11233-11433;\
,,11435-11467;,,11469-11491;,,11493-11641;,,11643-11662;,,11664-11706;,,11708-15910;\
,,15912-16044;,,16046-16080;,,16082-16148;,,16150-16291;,,16293-16336;,,16338-16405;\
,,16407-16474;,,16476-16908;,,16910-16915;,,16917-16944;,,16946-17155;,,17157-17187;\
,,17189-17389;,,17391-17423;,,17425-17447;,,17449-17597;,,17599-17618;,,17620-17662;,,17664-;";
cp new/tmpicosframe.pdb new/figs/fig13/figure_13_panel_B_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig12/figure_12_panel_H_2of3-1TMF_VP3_TRIMER.pdb new/figs/fig13/figure_13_panel_B_2of3-1TMF_VP3_TRIMER.pdb;
stripPDB --input=new/figs/fig12/figure_12_panel_H_2of3-1TMF_VP3_TRIMER.pdb \
--output=new/figs/fig13/figure_13_panel_B_3of3-1TMF_VP3_TRIMER_PROLINES.pdb \
--remove="\
,,-4245;,,4247-4260;,,4262-4380;,,4382-4416;,,4418-4481;,,4483-4618;,,4620-4663;,,4665-4733;\
,,4735-4802;,,4804-5233;,,5235-5240;,,5242-5269;,,5271-5482;,,5484-5514;,,5516-5608;\
,,5610-5735;,,5737-5769;,,5771-5793;,,5795-5943;,,5945-5964;,,5966-6008;,,6010-10508;\
,,10510-10523;,,10525-10643;,,10645-10679;,,10681-10744;,,10746-10881;,,10883-10926;\
,,10928-10996;,,10998-11065;,,11067-11496;,,11499-11503;,,11505-11532;,,11534-11745;\
,,11747-11777;,,11779-11871;,,11873-11998;,,12000-12032;,,12034-12056;,,12058-12206;\
,,12208-12227;,,12229-12271;,,12273-16771;,,16773-16786;,,16788-16906;,,16908-16942;\
,,16944-17007;,,17009-17144;,,17146-17189;,,17191-17259;,,17261-17328;,,17330-17759;\
,,17761-17766;,,17768-17795;,,17797-18008;,,18010-18040;,,18042-18134;,,18136-18261;\
,,18263-18295;,,18297-18319;,,18321-18469;,,18471-18490;,,18492-18534;,,18536-;";
cp new/tmpicosframe.pdb new/figs/fig13/figure_13_panels_C_1of5-ICOSAHEDRON.pdb;
cp new/figs/fig13/figure_13_panel_A_2of3-1TME_VP3_TRIMER.pdb new/figs/fig13/figure_13_panels_C_2of5-1TME_VP3_TRIMER.pdb;
cp new/figs/fig13/figure_13_panel_B_2of3-1TMF_VP3_TRIMER.pdb new/figs/fig13/figure_13_panels_C_3of5-1TMF_VP3_TRIMER.pdb;
cp new/figs/fig13/figure_13_panel_A_3of3-1TME_VP3_TRIMER_PROLINES.pdb new/figs/fig13/figure_13_panels_C_4of5-1TME_VP3_TRIMER_PROLINES.pdb;
cp new/figs/fig13/figure_13_panel_B_3of3-1TMF_VP3_TRIMER_PROLINES.pdb new/figs/fig13/figure_13_panels_C_5of5-1TME_VP3_TRIMER_PROLINES.pdb;
cp new/tmpicosframe.pdb new/figs/fig13/figure_13_panel_D_1of3-ICOSAHEDRON.pdb;
stripPDB --input=new/figs/fig13/figure_13_panel_A_3of3-1TME_VP3_TRIMER_PROLINES.pdb \
--output=new/figs/fig13/figure_13_panel_D_2of3-1TME_VP3_MONOMER_PROLINES.pdb \
--remove=",,5752-;";
stripPDB --input=new/figs/fig13/figure_13_panel_B_3of3-1TMF_VP3_TRIMER_PROLINES.pdb \
--output=new/figs/fig13/figure_13_panel_D_3of3-1TMF_VP3_MONOMER_PROLINES.pdb \
--remove=",,6010-;";
# Figure 14. Using Insert Residue Positions To Isolate Variations
# in Relative Position of Aligned Structures.
#
# Atoms are stripped from VP1234 trimers to make VP1-, VP2-, VP3-, and VP123- trimer substructures
# showing regions corresponding to Insert Residue Sections.
#
# The following is useful for panels in columns 2 - 6:
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
# Strip atoms to make Figure 14 VP1 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: TTSAGESADPVTTTVENYGG [1 - 20] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: ETQI [21 - 24] MP2 HELIX ONE
# VP1 I123-1: Q [25] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: RR [26 - 27] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHTDVSFIMDR [28 - 38] MP2 HELIX TWO
# VP1 M-5: FVKVTPQNQI [39 - 48] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: N [49] INSERTION
# VP1 B-1: IL [50 - 51] ABT CHAIN B
# VP1 I1-1: INSERTION
# VP1 B-2: ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: ABT CHAIN B
# VP1 I1-2: D [52] INSERTION
# VP1 B-4: LM [53 - 54] ABT CHAIN B
# VP1 I123-3: QVPSHT [55 - 60] INSERTION
# VP1 I1-3: LV [61 - 62] INSERTION
# VP1 I123-4: GG [63 - 64] INSERTION
# VP1 B-5: LLRAS [65 - 69] ABT CHAIN B
# VP1 M-6: TYYFSDLEIAVKH [70 - 82] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: EG [83 - 84] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: D [85] MP2 NON-HELIX
# VP1 I123-6: LT [86 - 87] INSERTION
# VP1 M-9: MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WV [88 - 89] MP2 NON-HELIX
# VP1 I13-2: PNGA [90 - 93] INSERTION
# VP1 I123-7: PE [94 - 95] INSERTION
# VP1 M-12: KALDNTTNPTAYHKAPLTR [96 - 114] MP2 NON-HELIX
# VP1 A-1: LALPYTAPHRVLATVYNGECRYSRNAV (PDB MISSING 'RYSRNAV') [115 - 141] ABT CHAIN A
# VP1 I1-4: PNLRGDLQVLAQ (PDB MISSING 'PNLRGDLQVLAQ') [142 - 153] INSERTION
# VP1 A-2: KVARTLPTSFNY (RES 154 - 156 'KVA' MISSING FROM PDB) [154 - 165] ABT CHAIN A
# VP1 M-13: GAIKAT [166 - 171] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: RVTELLYRMKRAETYCPRPLLA [172 - 193] MP2 NON-HELIX
# VP1 A-3: I [194] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: H [195] ABT CHAIN A
# VP1 M-15: PTEA [196 - 199] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RHKQKIVAPVK (PDB MISSING 'VK') [200 - 210] MP2 NON-HELIX
# VP1 A-6: QTL (PDB MISSING 'QTL') [211 - 213] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_A1_B1_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1BBT_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_A1_B1_2of5-1BBT_VP1_TRIMER.pdb \
--remove="BFJCGKDHL";
# VP1 I1-1: INSERTION
# VP1 I1-2: D [52] INSERTION
# VP1 I1-3: LV [61 - 62] INSERTION
# VP1 I1-4: PNLRGDLQVLAQ (PDB MISSING 'PNLRGDLQVLAQ') [142 - 153] INSERTION
# VP1 I1-5: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A1_B1_2of5-1BBT_VP1_TRIMER.pdb \
--remove=",-51;,53-60;,63-141;,154-;" \
--output=new/figs/fig14/figure_14_panels_A1_B1_3of5-INSERT_I1_OF_1BBT_VP1_TRIMER.pdb;
# VP1 I123-1: Q [25] INSERTION
# VP1 I123-2: INSERTION
# VP1 I123-3: QVPSHT [55 - 60] INSERTION
# VP1 I123-4: GG [63 - 64] INSERTION
# VP1 I123-5: INSERTION
# VP1 I123-6: LT [86 - 87] INSERTION
# VP1 I123-3: INSERTION
# VP1 I123-7: PE [94 - 95] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A1_B1_2of5-1BBT_VP1_TRIMER.pdb \
--remove=",-24;,26-54;,61-62;,65-85;,88-93;,96-;" \
--output=new/figs/fig14/figure_14_panels_A1_B1_5of5-INSERT_I123_OF_1BBT_VP1_TRIMER.pdb;
# VP1 I13-1: N [49] INSERTION
# VP1 I13-2: PNGA [90 - 93] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A1_B1_2of5-1BBT_VP1_TRIMER.pdb \
--remove=",-48;,50-89;,94-;" \
--output=new/figs/fig14/figure_14_panels_A1_B1_4of5-INSERT_I13_OF_1BBT_VP1_TRIMER.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GVDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FNY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDYRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSSSGPQKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGVN [99 - 102] INSERTION
# VP1 I123-4: GAN [103 - 105] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [106 - 120] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 I123-6: ASVL [142 - 145] INSERTION
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [147 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGA [149 - 152] INSERTION
# VP1 I123-7: PAD [153 - 155] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [156 - 176] MP2 NON-HELIX
# VP1 A-1: LVGAGNSQVSFVVPYNSPLSVLPAAWFNGWS [177 - 207] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [209 - 220] INSERTION
# VP1 A-2: ADFGRLWI [221 - 228] ABT CHAIN A
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [251 - 256] MP2 NON-HELIX
# VP1 I2-6: PT [257 - 258] INSERTION
# VP1 M-16: PTTTKINADNPVPILELE [259 - 276] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_A2_B2_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_A2_B2_2of5-1TMF_VP1_TRIMER.pdb \
--remove="BFJCGKDHL";
# VP1 I1-1: QENDYRLN [59 - 66] INSERTION
# VP1 I1-2: PDSSSGPQKTKA [78 - 89] INSERTION
# VP1 I1-3: GGVN [99 - 102] INSERTION
# VP1 I1-4: DFGNTKDFGVAPN [209 - 220] INSERTION
# VP1 I1-5: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A2_B2_2of5-1TMF_VP1_TRIMER.pdb \
--remove=",-58;,67-77;,90-98;,103-208;,221-;" \
--output=new/figs/fig14/figure_14_panels_A2_B2_3of5-INSERT_I1_OF_1TMF_VP1_TRIMER.pdb;
# VP1 I123-1: E [22] INSERTION
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I123-4: GAN [103 - 105] INSERTION
# VP1 I123-5: INSERTION
# VP1 I123-6: ASVL [142 - 145] INSERTION
# VP1 I123-7: PAD [153 - 155] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A2_B2_2of5-1TMF_VP1_TRIMER.pdb \
--remove=",-21;,23-26;,29-95;,99-102;,106-141;,146-152;,156-;" \
--output=new/figs/fig14/figure_14_panels_A2_B2_5of5-INSERT_I123_OF_1TMF_VP1_TRIMER.pdb;
# VP1 I13-1: FNY [56 - 58] INSERTION
# VP1 I13-2: PTGA [149 - 152] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A2_B2_2of5-1TMF_VP1_TRIMER.pdb \
--remove=",-55;,59-148;,153-;" \
--output=new/figs/fig14/figure_14_panels_A2_B2_4of5-INSERT_I13_OF_1TMF_VP1_TRIMER.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GSDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNIEST [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FVY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDLRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSTSGPVKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGT [99 - 101] INSERTION
# VP1 I123-4: TN [102 - 103] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [104 - 118] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [119 - 134] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [135 - 138] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [139] MP2 NON-HELIX
# VP1 I123-6: ASVL [140 - 143] INSERTION
# VP1 M-9: R [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [145 - 146] MP2 NON-HELIX
# VP1 I13-2: PTGA [147 - 150] INSERTION
# VP1 I123-7: PAD [151 - 153] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [154 - 174] MP2 NON-HELIX
# VP1 A-1: LVGAGNTQISFVVPYNSPLSVLPAAWFNGWS [175 - 205] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [206 - 218] INSERTION
# VP1 A-2: ADFGRLWI [219 - 226] ABT CHAIN A
# VP1 M-13: QGNTSA [227 - 232] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [233 - 248] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [249 - 254] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: PVSTRSKINADNPVPILELE [255 - 274] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_A3_B3_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_A3_B3_2of5-1TME_VP1_TRIMER.pdb \
--remove="BFJCGKDHL";
# VP1 I1-1: QENDLRLN [59 - 66] INSERTION
# VP1 I1-2: PDSTSGPVKTKA [78 - 89] INSERTION
# VP1 I1-3: GGT [99 - 101] INSERTION
# VP1 I1-4: DFGNTKDFGVAPN [206 - 218] INSERTION
# VP1 I1-5: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A3_B3_2of5-1TME_VP1_TRIMER.pdb \
--remove=",-58;,67-77;,90-98;,102-205;,219-;" \
--output=new/figs/fig14/figure_14_panels_A3_B3_3of5-INSERT_I1_OF_1TME_VP1_TRIMER.pdb;
# VP1 I123-1: E [22] INSERTION
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I123-4: TN [102 - 103] INSERTION
# VP1 I123-5: INSERTION
# VP1 I123-6: ASVL [140 - 143] INSERTION
# VP1 I123-7: PAD [151 - 153] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A3_B3_2of5-1TME_VP1_TRIMER.pdb \
--remove=",-21;,23-26;,29-95;,99-101;,104-139;,144-150;,154-;" \
--output=new/figs/fig14/figure_14_panels_A3_B3_5of5-INSERT_I123_OF_1TME_VP1_TRIMER.pdb;
# VP1 I13-1: FVY [56 - 58] INSERTION
# VP1 I13-2: PTGA [147 - 150] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A3_B3_2of5-1TME_VP1_TRIMER.pdb \
--remove=",-55;,59-146;,151-;" \
--output=new/figs/fig14/figure_14_panels_A3_B3_4of5-INSERT_I13_OF_1TME_VP1_TRIMER.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PALTAVETGATNPLV [43 - 56] MP2 NON-HELIX
# VP1 I3-1: P [57] INSERTION
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 I123-1: Q [62] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHRSRS [68 - 73] MP2 HELIX TWO
# VP1 M-5: ESSIESFFARGACVTIMT [74 - 91] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: V [92] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: DN [93 - 94] INSERTION
# VP1 B-2: PAS [95 - 97] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: TT [98 - 99] ABT CHAIN B
# VP1 I1-2: NKDK [100 - 103] INSERTION
# VP1 B-4: LFAVW [104 - 108] ABT CHAIN B
# VP1 I123-3: K [109] INSERTION
# VP1 I1-3: IT [110 - 111] INSERTION
# VP1 I123-4: YK [112 - 113] INSERTION
# VP1 B-5: DTVQLRRKLEF [114 - 124] ABT CHAIN B
# VP1 M-6: FTYSRFDMELTFVVTA [125 - 140] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NFTETNN [141 - 147] MP2 NON-HELIX
# VP1 I123-5: GHA [148 - 150] INSERTION
# VP1 M-8: LNQ [151 - 153] MP2 NON-HELIX
# VP1 I123-6: VY [154 - 155] INSERTION
# VP1 M-9: Q [156] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: IMYV [157 - 160] MP2 NON-HELIX
# VP1 I13-2: PPGA [161 - 164] INSERTION
# VP1 I123-7: PVPE [165 - 168] INSERTION
# VP1 M-12: KWDDYTWQTSSNPSIFYTYGTAPAR [169 - 193] MP2 NON-HELIX
# VP1 A-1: ISVPYVGISNAYSHFYDGFSKV [194 - 215] ABT CHAIN A
# VP1 I1-4: PLKDQSAALGDSLYGAASLN [216 - 235] INSERTION
# VP1 A-2: DFGILAVRV [236 - 244] ABT CHAIN A
# VP1 M-13: VNDHNPT [245 - 251] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: KVTSKIRVYLKPKHIRVWCPRPPRAVA [252 - 278] MP2 NON-HELIX
# VP1 A-3: YYG [279 - 281] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: PGVDY [282 - 286] ABT CHAIN A
# VP1 I1-5: KD [287 - 288] INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: GTLTP [289 - 293] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: LSTKD [294 - 298] MP2 NON-HELIX
# VP1 A-6: LTTY [299 - 302] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_A4_B4_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1HXS_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_A4_B4_2of5-1HXS_VP1_TRIMER.pdb \
--remove="BFJCGKDHL";
# VP1 I1-1: DN [93 - 94] INSERTION
# VP1 I1-2: NKDK [100 - 103] INSERTION
# VP1 I1-3: IT [110 - 111] INSERTION
# VP1 I1-4: PLKDQSAALGDSLYGAASLN [216 - 235] INSERTION
# VP1 I1-5: KD [287 - 288] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A4_B4_2of5-1HXS_VP1_TRIMER.pdb \
--remove=",-92;,95-99;,104-109;,112-215;,236-286;,289-;" \
--output=new/figs/fig14/figure_14_panels_A4_B4_3of5-INSERT_I1_OF_1HXS_VP1_TRIMER.pdb;
# VP1 I123-1: Q [62] INSERTION
# VP1 I123-2: INSERTION
# VP1 I123-3: K [109] INSERTION
# VP1 I123-4: YK [112 - 113] INSERTION
# VP1 I123-5: GHA [148 - 150] INSERTION
# VP1 I123-6: VY [154 - 155] INSERTION
# VP1 I123-7: PVPE [165 - 168] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A4_B4_2of5-1HXS_VP1_TRIMER.pdb \
--remove=",-61;,63-108;,110-111;,114-147;,151-153;,156-164;,169-;F;G;H;" \
--output=new/figs/fig14/figure_14_panels_A4_B4_5of5-INSERT_I123_OF_1HXS_VP1_TRIMER.pdb;
# VP1 I13-1: V [92] INSERTION
# VP1 I13-2: PPGA [161 - 164] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A4_B4_2of5-1HXS_VP1_TRIMER.pdb \
--remove=",-91;,93-160;,165-;" \
--output=new/figs/fig14/figure_14_panels_A4_B4_4of5-INSERT_I13_OF_1HXS_VP1_TRIMER.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_A5_B5_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_A5_B5_2of5-4WM7_VP1_TRIMER.pdb \
--remove="BFJCGKDHL";
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 I1-3: INT [95 -97] INSERTION
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 I1-5: KGRD [268 - 271] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A5_B5_2of5-4WM7_VP1_TRIMER.pdb \
--remove=",-76;,79-84;,89-94;,98-201;,215-267;,272-;" \
--output=new/figs/fig14/figure_14_panels_A5_B5_3of5-INSERT_I1_OF_4WM7_VP1_TRIMER.pdb;
# VP1 I123-1: Q [44] INSERTION
# VP1 I123-2: INSERTION
# VP1 I123-3: T [94] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A5_B5_2of5-4WM7_VP1_TRIMER.pdb \
--remove=",-43;,45-93;,95-97;,99-134;,139-140;,144-153;,156-;" \
--output=new/figs/fig14/figure_14_panels_A5_B5_5of5-INSERT_I123_OF_4WM7_VP1_TRIMER.pdb;
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 I13-2: PTGAL [149 - 153] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_A5_B5_2of5-4WM7_VP1_TRIMER.pdb \
--remove=",-73;,77-148;,154-;" \
--output=new/figs/fig14/figure_14_panels_A5_B5_4of5-INSERT_I13_OF_4WM7_VP1_TRIMER.pdb;
# Strip atoms to make Figure 14 VP2 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: DKK (RES 1-3 'DKK' MISSING FROM PDB) [1 - 3] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: TEETTLL (RES 4-8 'TEETT' MISSING FROM PDB) [4 - 10] MP2 HELIX ONE
# VP2 I123-1: E [11] INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: D [12] INSERTION
# VP2 M-3: RI [13 - 14] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LTTRNGH [15 - 21] MP2 HELIX TWO
# VP2 M-5: TTSTTQSSVG [22 - 31] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: VTYG [32 - 35] ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: YATA [36 - 39] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: E [40] ABT CHAIN B
# VP2 I123-3: INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: INSERTION
# VP2 B-5: DFVSGP [41 - 46] ABT CHAIN B
# VP2 M-6: NTSGLETRVV [47 - 56] MP2 NON-HELIX
# VP2 I2-1: QA [57 - 58] INSERTION
# VP2 I23-2: ERFFKTHLFDW [59 - 69] INSERTION
# VP2 M-7: VTSDS [70 - 74] MP2 NON-HELIX
# VP2 I123-5: FGR [75 - 77] INSERTION
# VP2 M-8: MP2 NON-HELIX
# VP2 I123-6: CHLL [78 - 81] INSERTION
# VP2 M-9: MP2 NON-HELIX
# VP2 I23-3: E [82] INSERTION
# VP2 I2-2: INSERTION
# VP2 M-10: MP2 NON-HELIX
# VP2 I2-3: INSERTION
# VP2 M-11: L [83] MP2 NON-HELIX
# VP2 I13-2: P [84] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: TDHKGVYGSLTDSYA [85 - 99] MP2 NON-HELIX
# VP2 A-1: YMRNGWDVEVTAV [100 - 112] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: GNQF [113 - 116] ABT CHAIN A
# VP2 M-13: NGGCLL [117 - 122] MP2 NON-HELIX
# VP2 I2-4: VAM [123 - 125] INSERTION
# VP2 M-14: VPELCSIQKRE [126 - 136] MP2 NON-HELIX
# VP2 A-3: ABT CHAIN A
# VP2 I2-5: LYQLTLFPHQFINPRTNMTAHITVPFVGVNR [137 - 167] INSERTION
# VP2 A-4: YDQY [168 - 171] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: KVHK [172 - 175] ABT CHAIN A
# VP2 M-15: PWTLVVMVVA [176 - 185] MP2 NON-HELIX
# VP2 I2-6: PLTVNTEGAPQIKVYANIAPTN [186 - 207] INSERTION
# VP2 M-16: VHVAGEFPSKE [208 - 218] MP2 NON-HELIX
# VP2 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_C1_D1_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1BBT_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_C1_D1_2of5-1BBT_VP2_TRIMER.pdb \
--remove="AEICGKDHL";
# VP2 I123-1: E [11] INSERTION
# VP2 I123-2: INSERTION
# VP2 I123-3: INSERTION
# VP2 I123-4: INSERTION
# VP2 I123-5: FGR [75 - 77] INSERTION
# VP2 I123-6: CHLL [78 - 81] INSERTION
# VP2 I123-7: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C1_D1_2of5-1BBT_VP2_TRIMER.pdb \
--remove=",-10;,12-74;,82-;" \
--output=new/figs/fig14/figure_14_panels_C1_D1_5of5-INSERT_I123_OF_1BBT_VP2_TRIMER.pdb;
# VP2 I2-1: QA [57 - 58] INSERTION
# VP2 I2-2: INSERTION
# VP2 I2-3: INSERTION
# VP2 I2-4: VAM [123 - 125] INSERTION
# VP2 I2-5: LYQLTLFPHQFINPRTNMTAHITVPFVGVNR [137 - 167] INSERTION
# VP2 I2-6: PLTVNTEGAPQIKVYANIAPTN [186 - 207] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C1_D1_2of5-1BBT_VP2_TRIMER.pdb \
--remove=",-56;,59-122;,126-136;,168-185;,208-;" \
--output=new/figs/fig14/figure_14_panels_C1_D1_3of5-INSERT_I2_OF_1BBT_VP2_TRIMER.pdb;
# VP2 I23-1: D [12] INSERTION
# VP2 I23-2: ERFFKTHLFDW [59 - 69] INSERTION
# VP2 I23-3: E [82] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C1_D1_2of5-1BBT_VP2_TRIMER.pdb \
--remove=",-11;,13-58;,70-81;,83-;" \
--output=new/figs/fig14/figure_14_panels_C1_D1_4of5-INSERT_I23_OF_1BBT_VP2_TRIMER.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: DQNTEEM [1 - 7] MP2 HELIX ONE
# VP2 I123-1: E [8] INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: NLSD [9 - 12] INSERTION
# VP2 M-3: RVA [13 - 15] MP2 HELIX ONE
# VP2 I123-2: SD [16 - 17] INSERTION
# VP2 M-4: KAG [18 - 20] MP2 HELIX TWO
# VP2 M-5: NSATNTQSTVGR [21 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: L [33] ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [34] ABT CHAIN B
# VP2 I3-4: G [35] INSERTION
# VP2 B-3: Y [36] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GK [37 - 38] ABT CHAIN B
# VP2 I123-3: S [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: HHG [40 - 42] INSERTION
# VP2 B-5: EHPASCA [43 - 49] ABT CHAIN B
# VP2 M-6: DTATDKVL [50 - 57] MP2 NON-HELIX
# VP2 I2-1: AA [58 - 59] INSERTION
# VP2 I23-2: ERYYTIDLASWTTS [60 - 73] INSERTION
# VP2 M-7: QEAFS [74 - 78] MP2 NON-HELIX
# VP2 I123-5: HIRIPLP [79 - 85] INSERTION
# VP2 M-8: MP2 NON-HELIX
# VP2 I123-6: HVLAG [86 - 90] INSERTION
# VP2 M-9: MP2 NON-HELIX
# VP2 I23-3: EDGG [91 - 94] INSERTION
# VP2 I2-2: VFGATLRRHYL [95 - 105] INSERTION
# VP2 M-10: CKTG [106 - 109] MP2 NON-HELIX
# VP2 I2-3: WRVQVQCNASQFHAGSLL [110 - 127] INSERTION
# VP2 M-11: VFMAP [128 - 132] MP2 NON-HELIX
# VP2 I13-2: INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EFYTGKGTKTGTMEP [133 - 147] MP2 NON-HELIX
# VP2 A-1: SDPFTMDTEWRS P [148 - 160] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: QGAPTGYRY [161 - 169] ABT CHAIN A
# VP2 M-13: DSRTGFF [170 - 176] MP2 NON-HELIX
# VP2 I2-4: ATN [177 - 179] INSERTION
# VP2 M-14: HQNQW [180 - 184] MP2 NON-HELIX
# VP2 A-3: ABT CHAIN A
# VP2 I2-5: QWTVYPHQILNLRTNTTVDLEVPYVNVAP [185 - 213] INSERTION
# VP2 A-4: SSSWTQ [214 - 219] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: H [220] ABT CHAIN A
# VP2 M-15: ANWTLVVAVLS [221 - 231] MP2 NON-HELIX
# VP2 I2-6: PLQYATGSSPDVQITASLQPVNPVFNG [232 - 258] INSERTION
# VP2 M-16: LRHETV [259 - 264] MP2 NON-HELIX
# VP2 A-6: IAQ [265 - 267] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_C2_D2_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_C2_D2_2of5-1TMF_VP2_TRIMER.pdb \
--remove="AEICGKDHL";
# VP2 I123-1: E [8] INSERTION
# VP2 I123-2: SD [16 - 17] INSERTION
# VP2 I123-3: S [39] INSERTION
# VP2 I123-4: HHG [40 - 42] INSERTION
# VP2 I123-5: HIRIPLP [79 - 85] INSERTION
# VP2 I123-6: HVLAG [86 - 90] INSERTION
# VP2 I123-7: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C2_D2_2of5-1TMF_VP2_TRIMER.pdb \
--remove=",-7;,9-15;,18-38;,43-78;,91-;" \
--output=new/figs/fig14/figure_14_panels_C2_D2_5of5-INSERT_I123_OF_1TMF_VP2_TRIMER.pdb;
# VP2 I2-1: AA [58 - 59] INSERTION
# VP2 I2-2: VFGATLRRHYL [95 - 105] INSERTION
# VP2 I2-3: WRVQVQCNASQFHAGSLL [110 - 127] INSERTION
# VP2 I2-4: ATN [177 - 179] INSERTION
# VP2 I2-5: QWTVYPHQILNLRTNTTVDLEVPYVNVAP [185 - 213] INSERTION
# VP2 I2-6: PLQYATGSSPDVQITASLQPVNPVFNG [232 - 258] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C2_D2_2of5-1TMF_VP2_TRIMER.pdb \
--remove=",-57;,60-94;,106-109;,128-176;,180-184;,214-231;,259-;" \
--output=new/figs/fig14/figure_14_panels_C2_D2_3of5-INSERT_I2_OF_1TMF_VP2_TRIMER.pdb;
# VP2 I23-1: NLSD [9 - 12] INSERTION
# VP2 I23-2: ERYYTIDLASWTTS [60 - 73] INSERTION
# VP2 I23-3: EDGG [91 - 94] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C2_D2_2of5-1TMF_VP2_TRIMER.pdb \
--remove=",-8;,13-59;,74-90;,95-;" \
--output=new/figs/fig14/figure_14_panels_C2_D2_4of5-INSERT_I23_OF_1TMF_VP2_TRIMER.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: DQNTEEM (RES 1 - 7 'DQNTTEM' MISSING FROM PDB) [1 - 7] MP2 HELIX ONE
# VP2 I123-1: E (RES 8 'E' MISSING FROM PDB) [8] INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: NLSD (RES 9 - 12 'NLS' MISSING FROM PDB) [9 - 12] INSERTION
# VP2 M-3: RVA [13 - 15] MP2 HELIX ONE
# VP2 I123-2: SD [16 - 17] INSERTION
# VP2 M-4: KAG [18 - 20] MP2 HELIX TWO
# VP2 M-5: NSATNTQSTVGR [21 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: L [33] ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [34] ABT CHAIN B
# VP2 I3-4: G [35] INSERTION
# VP2 B-3: Y [36] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GEA [37 - 39] ABT CHAIN B
# VP2 I123-3: INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: HHG [40 - 42] INSERTION
# VP2 B-5: EHPASCA [43 - 49] ABT CHAIN B
# VP2 M-6: DTATDKVL [50 - 57] MP2 NON-HELIX
# VP2 I2-1: AA [58 - 59] INSERTION
# VP2 I23-2: ERYYTIDLASWTTT [60 - 73] INSERTION
# VP2 M-7: QEAFS [74 - 78] MP2 NON-HELIX
# VP2 I123-5: HIRIPLP [79 - 85] INSERTION
# VP2 M-8: MP2 NON-HELIX
# VP2 I123-6: HVLAG [86 - 90] INSERTION
# VP2 M-9: MP2 NON-HELIX
# VP2 I23-3: EDGG [91 - 94] INSERTION
# VP2 I2-2: VFGATLRRHYL [95 - 105] INSERTION
# VP2 M-10: CKTG [106 - 109] MP2 NON-HELIX
# VP2 I2-3: WRVQVQCNASQFHAGSLL [110 - 127] INSERTION
# VP2 M-11: VFMAP [128 - 132] MP2 NON-HELIX
# VP2 I13-2: INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EFYTGKGTKTGDMEP [133 - 147] MP2 NON-HELIX
# VP2 A-1: TDPFTMDTTWRAP [148 - 160] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: QGAPTGYRY [161 - 169] ABT CHAIN A
# VP2 M-13: DSRTGFF [170 - 176] MP2 NON-HELIX
# VP2 I2-4: AMN [177 - 179] INSERTION
# VP2 M-14: HQNQW [180 - 184] MP2 NON-HELIX
# VP2 A-3: ABT CHAIN A
# VP2 I2-5: QWTVYPHQILNLRTNTTVDLEVPYVNIAP [185 - 213] INSERTION
# VP2 A-4: TSSWTQ [214 - 219] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: H [220] ABT CHAIN A
# VP2 M-15: ANWTLVVAVFS [221 - 231] MP2 NON-HELIX
# VP2 I2-6: PLQYASGSSSDVQITASIQPVNPVFNG [232 - 258] INSERTION
# VP2 M-16: LRHETV [259 - 264] MP2 NON-HELIX
# VP2 A-6: IAQ (RES 267 'Q' MISSING FROM PDB) [265 - 267] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_C3_D3_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_C3_D3_2of5-1TME_VP2_TRIMER.pdb \
--remove="AEICGKDHL";
# VP2 I123-1: E (RES 8 'E' MISSING FROM PDB) [8] INSERTION
# VP2 I123-2: SD [16 - 17] INSERTION
# VP2 I123-3: INSERTION
# VP2 I123-4: HHG [40 - 42] INSERTION
# VP2 I123-5: HIRIPLP [79 - 85] INSERTION
# VP2 I123-6: HVLAG [86 - 90] INSERTION
# VP2 I123-7: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C3_D3_2of5-1TME_VP2_TRIMER.pdb \
--remove=",-7;,9-15;,18-39;,43-78;,91-;" \
--output=new/figs/fig14/figure_14_panels_C3_D3_5of5-INSERT_I123_OF_1TME_VP2_TRIMER.pdb;
# VP2 I2-1: AA [58 - 59] INSERTION
# VP2 I2-2: VFGATLRRHYL [95 - 105] INSERTION
# VP2 I2-3: WRVQVQCNASQFHAGSLL [110 - 127] INSERTION
# VP2 I2-4: AMN [177 - 179] INSERTION
# VP2 I2-5: QWTVYPHQILNLRTNTTVDLEVPYVNIAP [185 - 213] INSERTION
# VP2 I2-6: PLQYASGSSSDVQITASIQPVNPVFNG [232 - 258] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C3_D3_2of5-1TME_VP2_TRIMER.pdb \
--remove=",-57;,60-94;,106-109;,128-176;,180-184;,214-231;,259-;" \
--output=new/figs/fig14/figure_14_panels_C3_D3_3of5-INSERT_I2_OF_1TME_VP2_TRIMER.pdb;
# VP2 I23-1: NLSD (RES 9 - 12 'NLS' MISSING FROM PDB) [9 - 12] INSERTION
# VP2 I23-2: ERYYTIDLASWTTT [60 - 73] INSERTION
# VP2 I23-3: EDGG [91 - 94] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C3_D3_2of5-1TME_VP2_TRIMER.pdb \
--remove=",-8;,13-59;,74-90;,95-;" \
--output=new/figs/fig14/figure_14_panels_C3_D3_4of5-INSERT_I23_OF_1TME_VP2_TRIMER.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: S (RES 1 'S' MISSING) [1] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: PNIEA (RES 2-5 'PNIE' MISSING) [2 - 6] MP2 HELIX ONE
# VP2 I123-1: INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: CGYSD [7 - 11] INSERTION
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LQLTLG [14 - 19] MP2 HELIX TWO
# VP2 M-5: NSTITTQEAANSV [20 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: V [33] ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: AY [34 - 35] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GRW [36 - 37] ABT CHAIN B
# VP2 I123-3: P [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: INSERTION
# VP2 B-5: EYLRDS [40 - 45] ABT CHAIN B
# VP2 M-6: EANPVDQ [46 - 52] MP2 NON-HELIX
# VP2 I2-1: PTEPDVAAC [53 - 61] INSERTION
# VP2 I23-2: RFYTLDTVSWT [62 - 72] INSERTION
# VP2 M-7: KES [73 - 75] MP2 NON-HELIX
# VP2 I123-5: RGWWWKLP [76 - 83] INSERTION
# VP2 M-8: D [84] MP2 NON-HELIX
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 M-9: R [87] MP2 NON-HELIX
# VP2 I23-3: DM [88 - 89] INSERTION
# VP2 I2-2: GLFGQNMYYHYLG [90 - 102] INSERTION
# VP2 M-10: RS [103 - 104] MP2 NON-HELIX
# VP2 I2-3: GYTVHVQCNASKFHQGALG [105 - 123] INSERTION
# VP2 M-11: VFAV [124 - 127] MP2 NON-HELIX
# VP2 I13-2: P [128] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EMCLAGDSNT [129 - 138] MP2 NON-HELIX
# VP2 A-1: TTMHTSYQNANPGEKGG [139 - 155] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: TFT [156 - 158] ABT CHAIN A
# VP2 M-13: GTFT [159 - 162] MP2 NON-HELIX
# VP2 I2-4: PDNN [163 - 166] INSERTION
# VP2 M-14: QTSPARRFCPVDYLL [167 - 181] MP2 NON-HELIX
# VP2 A-3: GNGT [182 - 185] ABT CHAIN A
# VP2 I2-5: LLGNAFVFPHQIINLRTNNCATLVLPYVNSLS [186 - 217] INSERTION
# VP2 A-4: IDSMV [218 - 222] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: KH [223 - 224] ABT CHAIN A
# VP2 M-15: NNWGIAILPLA [225 - 235] MP2 NON-HELIX
# VP2 I2-6: PLNFASESSPEIPITLTIAPMCCEFNG [236 - 262] INSERTION
# VP2 M-16: LRNITLP [263 - 269] MP2 NON-HELIX
# VP2 A-6: RLQ [270 - 272] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_C4_D4_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1HXS_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_C4_D4_2of5-1HXS_VP2_TRIMER.pdb \
--remove="AEICGKDHL";
# VP2 I123-1: INSERTION
# VP2 I123-2: INSERTION
# VP2 I123-3: P [39] INSERTION
# VP2 I123-4: INSERTION
# VP2 I123-5: RGWWWKLP [76 - 83] INSERTION
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 I123-7: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C4_D4_2of5-1HXS_VP2_TRIMER.pdb \
--remove=",-38;,40-75;,84;,87-;" \
--output=new/figs/fig14/figure_14_panels_C4_D4_5of5-INSERT_I123_OF_1HXS_VP2_TRIMER.pdb;
# VP2 I2-1: PTEPDVAAC [53 - 61] INSERTION
# VP2 I2-2: GLFGQNMYYHYLG [90 - 102] INSERTION
# VP2 I2-3: GYTVHVQCNASKFHQGALG [105 - 123] INSERTION
# VP2 I2-4: PDNN [163 - 166] INSERTION
# VP2 I2-5: LLGNAFVFPHQIINLRTNNCATLVLPYVNSLS [186 - 217] INSERTION
# VP2 I2-6: PLNFASESSPEIPITLTIAPMCCEFNG [236 - 262] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C4_D4_2of5-1HXS_VP2_TRIMER.pdb \
--remove=",-52;,62-89;,103-104;,124-162;,167-185;,218-235;,263-;" \
--output=new/figs/fig14/figure_14_panels_C4_D4_3of5-INSERT_I2_OF_1HXS_VP2_TRIMER.pdb;
# VP2 I23-1: CGYSD [7 - 11] INSERTION
# VP2 I23-2: RFYTLDTVSWT [62 - 72] INSERTION
# VP2 I23-3: DM [88 - 89] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C4_D4_2of5-1HXS_VP2_TRIMER.pdb \
--remove=",-6;,12-61;,73-87;,90-;" \
--output=new/figs/fig14/figure_14_panels_C4_D4_4of5-INSERT_I23_OF_1HXS_VP2_TRIMER.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP2 M-1: S (RES 1 'S' MISSING FROM PDB) [1] MP2 NON-HELIX
# VP2 I3-1: INSERTION
# VP2 M-2: PSAEA (RES 2-6 'PSAEA' MISSING FROM PDB) [2 - 6] MP2 HELIX ONE
# VP2 I123-1: INSERTION
# VP2 I3-2: INSERTION
# VP2 I23-1: CGYSD (RES 7-9 'CGY' MISSING FROM PDB) [7 - 11] INSERTION
# VP2 M-3: RV [12 - 13] MP2 HELIX ONE
# VP2 I123-2: INSERTION
# VP2 M-4: LQLKLG [14 - 19] MP2 HELIX TWO
# VP2 M-5: NSAIVTQEAANYC [20 - 32] MP2 NON-HELIX
# VP2 I3-3: INSERTION
# VP2 I13-1: INSERTION
# VP2 B-1: ABT CHAIN B
# VP2 I1-1: INSERTION
# VP2 B-2: C [33] ABT CHAIN B
# VP2 I3-4: INSERTION
# VP2 B-3: AY [34 - 35] ABT CHAIN B
# VP2 I1-2: INSERTION
# VP2 B-4: GEW [36 - 38] ABT CHAIN B
# VP2 I123-3: P [39] INSERTION
# VP2 I1-3: INSERTION
# VP2 I123-4: N [40] INSERTION
# VP2 B-5: YLPD [41 - 44] ABT CHAIN B
# VP2 M-6: HEAVAIDK [45 - 52] MP2 NON-HELIX
# VP2 I2-1: PTQPETST [53 - 60] INSERTION
# VP2 I23-2: DRFYTLRSVKW [61 - 71] INSERTION
# VP2 M-7: ESNST [72 - 76] MP2 NON-HELIX
# VP2 I123-5: GWWWKL [77 - 82] INSERTION
# VP2 M-8: PD [83 - 84] MP2 NON-HELIX
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 M-9: NN [87 - 88] MP2 NON-HELIX
# VP2 I23-3: IGMFGQNVQYHYLY [89 - 102] INSERTION
# VP2 I2-2: RS [103 - 104] INSERTION
# VP2 M-10: GFLIHVQCNATKFHQGALL [105 - 123] MP2 NON-HELIX
# VP2 I2-3: INSERTION
# VP2 M-11: VVAI [124 - 127] MP2 NON-HELIX
# VP2 I13-2: P [128] INSERTION
# VP2 I123-7: INSERTION
# VP2 M-12: EHQRGAHDTT [129 - 138] MP2 NON-HELIX
# VP2 A-1: TSPGF [139 - 143] ABT CHAIN A
# VP2 I1-4: INSERTION
# VP2 A-2: NDIMK [144 - 148] ABT CHAIN A
# VP2 M-13: GERGGTF [149 - 155] MP2 NON-HELIX
# VP2 I2-4: N [156] INSERTION
# VP2 M-14: HPYVL [157 - 161] MP2 NON-HELIX
# VP2 A-3: DDGTS [162 - 166] ABT CHAIN A
# VP2 I2-5: IACATIFPHQWINLRTNNSATIVLPWMNVAP [167 - 197] INSERTION
# VP2 A-4: MDFPL [198 - 202] ABT CHAIN A
# VP2 I1-5: INSERTION
# VP2 A-5: RH [203 - 204] ABT CHAIN A
# VP2 M-15: NQWTLAVIPVV [205 - 215] MP2 NON-HELIX
# VP2 I2-6: PLGTRTMSSVVPITVSIAPMCCEFNG [216 - 241] INSERTION
# VP2 M-16: LRHAIT [242 - 247] MP2 NON-HELIX
# VP2 A-6: Q [248] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_C5_D5_1of5-ICOSAHEDRON.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_C5_D5_2of5-4WM7_VP2_TRIMER.pdb \
--remove="AEICGKDHL";
# VP2 I123-1: INSERTION
# VP2 I123-2: INSERTION
# VP2 I123-3: P [39] INSERTION
# VP2 I123-4: N [40] INSERTION
# VP2 I123-5: GWWWKL [77 - 82] INSERTION
# VP2 I123-6: AL [85 - 86] INSERTION
# VP2 I123-7: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C5_D5_2of5-4WM7_VP2_TRIMER.pdb \
--remove=",-38;,41-76;,83-84;,87-;" \
--output=new/figs/fig14/figure_14_panels_C5_D5_5of5-INSERT_I123_OF_4WM7_VP2_TRIMER.pdb;
# VP2 I2-1: PTQPETST [53 - 60] INSERTION
# VP2 I2-2: RS [103 - 104] INSERTION
# VP2 I2-3: INSERTION
# VP2 I2-4: N [156] INSERTION
# VP2 I2-5: IACATIFPHQWINLRTNNSATIVLPWMNVAP [167 - 197] INSERTION
# VP2 I2-6: PLGTRTMSSVVPITVSIAPMCCEFNG [216 - 241] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C5_D5_2of5-4WM7_VP2_TRIMER.pdb \
--remove=",-52;,61-102;,105-155;,157-166;,198-215;,242-;" \
--output=new/figs/fig14/figure_14_panels_C5_D5_3of5-INSERT_I2_OF_4WM7_VP2_TRIMER.pdb;
# VP2 I23-1: CGYSD (RES 7-9 'CGY' MISSING FROM PDB) [7 - 11] INSERTION
# VP2 I23-2: DRFYTLRSVKW [61 - 71] INSERTION
# VP2 I23-3: IGMFGQNVQYHYLY [89 - 102] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_C5_D5_2of5-4WM7_VP2_TRIMER.pdb \
--remove=",-6;,12-60;,72-88;,103-;" \
--output=new/figs/fig14/figure_14_panels_C5_D5_4of5-INSERT_I23_OF_4WM7_VP2_TRIMER.pdb;
# Strip atoms to make Figure 14 VP3 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: ACPTFLRFEGGV [50 - 61] MP2 NON-HELIX
# VP3 I3-1: PYVTTK [62 - 67] INSERTION
# VP3 M-2: TDSD [68 - 71] MP2 HELIX ONE
# VP3 I123-1: INSERTION
# VP3 I3-2: RV [72 - 73] INSERTION
# VP3 I23-1: LAQFD [74 - 78] INSERTION
# VP3 M-3: MP2 HELIX ONE
# VP3 I123-2: MS [79 - 80] INSERTION
# VP3 M-4: LAAKHMSNTFLAG [81 - 93] MP2 HELIX TWO
# VP3 M-5: LAQYYT [94 - 99] MP2 NON-HELIX
# VP3 I3-3: QYS [100 - 102] INSERTION
# VP3 I13-1: GTIN [103 - 106] INSERTION
# VP3 B-1: L [107] ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: HF [108 - 109] ABT CHAIN B
# VP3 I3-4: MFTGPTDAKARYMV [110 - 123] INSERTION
# VP3 B-3: AYA [124 - 126] ABT CHAIN B
# VP3 I1-2: P [127] INSERTION
# VP3 B-4: PGME [128 - 131] ABT CHAIN B
# VP3 I123-3: P [132] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: PKT [133 - 135] INSERTION
# VP3 B-5: PEAAAHCI [136 - 143] ABT CHAIN B
# VP3 M-6: HAEWDTGLNSKFTFSIPYLSAA [144 - 165] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: DYTYTAS [166 - 172] INSERTION
# VP3 M-7: DVAETT [173 - 178] MP2 NON-HELIX
# VP3 I123-5: NVQGWVCLF [179 - 187] INSERTION
# VP3 M-8: Q [188] MP2 NON-HELIX
# VP3 I123-6: INSERTION
# VP3 M-9: MP2 NON-HELIX
# VP3 I23-3: INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: I [189] MP2 NON-HELIX
# VP3 I13-2: INSERTION
# VP3 I123-7: TH [190 - 191] INSERTION
# VP3 M-12: GKADGDALVVLASAGK [192 - 207] MP2 NON-HELIX
# VP3 A-1: DFELRLPV [208 - 215] ABT CHAIN A
# VP3 I1-4: DAR [216 - 218] INSERTION
# VP3 A-2: ABT CHAIN A
# VP3 M-13: AE [219 - 220] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_E1_F1_1of6-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1BBT_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_E1_F1_2of6-1BBT_VP3_TRIMER.pdb \
--remove="AEIBFJDHL";
# VP3 I123-1: INSERTION
# VP3 I123-2: MS [79 - 80] INSERTION
# VP3 I123-3: P [132] INSERTION
# VP3 I123-4: PKT [133 - 135] INSERTION
# VP3 I123-5: NVQGWVCLF [179 - 187] INSERTION
# VP3 I123-6: INSERTION
# VP3 I123-7: TH [190 - 191] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E1_F1_2of6-1BBT_VP3_TRIMER.pdb \
--remove=",-78;,81-131;,136-178;,188-189;,192-;" \
--output=new/figs/fig14/figure_14_panels_E1_F1_6of6-INSERT_I123_OF_1BBT_VP3_TRIMER.pdb;
# VP3 I13-1: GTIN [103 - 106] INSERTION
# VP3 I13-2: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E1_F1_2of6-1BBT_VP3_TRIMER.pdb \
--remove=",-102;,107-;" \
--output=new/figs/fig14/figure_14_panels_E1_F1_4of6-INSERT_I13_OF_1BBT_VP3_TRIMER.pdb;
# VP3 I23-1: LAQFD [74 - 78] INSERTION
# VP3 I23-2: DYTYTAS [166 - 172] INSERTION
# VP3 I23-3: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E1_F1_2of6-1BBT_VP3_TRIMER.pdb \
--remove=",-73;,79-165;,173-;" \
--output=new/figs/fig14/figure_14_panels_E1_F1_5of6-INSERT_I23_OF_1BBT_VP3_TRIMER.pdb;
# VP3 I3-1: PYVTTK [62 - 67] INSERTION
# VP3 I3-2: RV [72 - 73] INSERTION
# VP3 I3-3: QYS [100 - 102] INSERTION
# VP3 I3-4: MFTGPTDAKARYMV [110 - 123] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E1_F1_2of6-1BBT_VP3_TRIMER.pdb \
--remove=",-61;,68-71;,74-99;,103-109;,124-;" \
--output=new/figs/fig14/figure_14_panels_E1_F1_3of6-INSERT_I3_OF_1BBT_VP3_TRIMER.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: PNTNNKRY [57 - 64] MP2 NON-HELIX
# VP3 I3-1: PYFSATNSV [65 - 73] INSERTION
# VP3 M-2: PATSMVDY [74 - 81] MP2 HELIX ONE
# VP3 I123-1: Q [82] INSERTION
# VP3 I3-2: VALSCS [83 - 88] INSERTION
# VP3 I23-1: CMAN [89 - 92] INSERTION
# VP3 M-3: S [93] MP2 HELIX ONE
# VP3 I123-2: M [94] INSERTION
# VP3 M-4: LAAVARN [95 - 101] MP2 HELIX TWO
# VP3 M-5: FNQYR [102 - 106] MP2 NON-HELIX
# VP3 I3-3: INSERTION
# VP3 I13-1: GSLN [107 - 110] INSERTION
# VP3 B-1: FL [111 - 112] ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: F [113] ABT CHAIN B
# VP3 I3-4: VFTGAAMVKGKFLI [114 - 127] INSERTION
# VP3 B-3: AYT [128 - 130] ABT CHAIN B
# VP3 I1-2: P [131] INSERTION
# VP3 B-4: PGAGK [132 - 136] ABT CHAIN B
# VP3 I123-3: P [137] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: T [138] INSERTION
# VP3 B-5: TRDQAM [139 - 144] ABT CHAIN B
# VP3 M-6: QSTYAIWDLGLNSSFNFTAPFISPTH [145 - 170] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRQTSYTSPTITS [171 - 183] INSERTION
# VP3 M-7: VD [184 - 185] MP2 NON-HELIX
# VP3 I123-5: GWVTVW [186 - 191] INSERTION
# VP3 M-8: Q [192] MP2 NON-HELIX
# VP3 I123-6: LT [193 - 194] INSERTION
# VP3 M-9: P [195] MP2 NON-HELIX
# VP3 I23-3: LT [196 - 197] INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: Y [198] MP2 NON-HELIX
# VP3 I13-2: PSG [199 - 201] INSERTION
# VP3 I123-7: TP [202 - 203] INSERTION
# VP3 M-12: TNSDILTLVSAG [204 - 215] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: DDFTLRMPISPTKWV [216 - 230] ABT CHAIN A
# VP3 M-13: P [231] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: Q [232] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_E2_F2_1of6-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_E2_F2_2of6-1TMF_VP3_TRIMER.pdb \
--remove="AEIBFJDHL";
# VP3 I123-1: Q [82] INSERTION
# VP3 I123-2: M [94] INSERTION
# VP3 I123-3: P [137] INSERTION
# VP3 I123-4: T [138] INSERTION
# VP3 I123-5: GWVTVW [186 - 191] INSERTION
# VP3 I123-6: LT [193 - 194] INSERTION
# VP3 I123-7: TP [202 - 203] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E2_F2_2of6-1TMF_VP3_TRIMER.pdb \
--remove=",-81;,83-93;,95-136;,139-185;,192;,195-201;,204-;" \
--output=new/figs/fig14/figure_14_panels_E2_F2_6of6-INSERT_I123_OF_1TMF_VP3_TRIMER.pdb;
# VP3 I13-1: GSLN [107 - 110] INSERTION
# VP3 I13-2: PSG [199 - 201] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E2_F2_2of6-1TMF_VP3_TRIMER.pdb \
--remove=",-106;,111-198;,202-;" \
--output=new/figs/fig14/figure_14_panels_E2_F2_4of6-INSERT_I13_OF_1TMF_VP3_TRIMER.pdb;
# VP3 I23-1: CMAN [89 - 92] INSERTION
# VP3 I23-2: YRQTSYTSPTITS [171 - 183] INSERTION
# VP3 I23-3: LT [196 - 197] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E2_F2_2of6-1TMF_VP3_TRIMER.pdb \
--remove=",-88;,93-170;,184-195;,198-;" \
--output=new/figs/fig14/figure_14_panels_E2_F2_5of6-INSERT_I23_OF_1TMF_VP3_TRIMER.pdb;
# VP3 I3-1: PYFSATNSV [65 - 73] INSERTION
# VP3 I3-2: VALSCS [83 - 88] INSERTION
# VP3 I3-3: INSERTION
# VP3 I3-4: VFTGAAMVKGKFLI [114 - 127] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E2_F2_2of6-1TMF_VP3_TRIMER.pdb \
--remove=",-64;,74-82;,89-113;,128-;" \
--output=new/figs/fig14/figure_14_panels_E2_F2_3of6-INSERT_I3_OF_1TMF_VP3_TRIMER.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: PNSNNKRY [57 - 64] MP2 NON-HELIX
# VP3 I3-1: PYFSATNSV [65 - 73] INSERTION
# VP3 M-2: PTTSLVDY [74 - 81] MP2 HELIX ONE
# VP3 I123-1: Q [82] INSERTION
# VP3 I3-2: VALSCS [83 - 88] INSERTION
# VP3 I23-1: CMCN [89 - 92] INSERTION
# VP3 M-3: S [93] MP2 HELIX ONE
# VP3 I123-2: M [94] INSERTION
# VP3 M-4: LAAVARN [95 - 101] MP2 HELIX TWO
# VP3 M-5: FNQYR [102 - 106] MP2 NON-HELIX
# VP3 I3-3: INSERTION
# VP3 I13-1: GSLN [107 - 110] INSERTION
# VP3 B-1: FL [111 - 112] ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: F [113] ABT CHAIN B
# VP3 I3-4: VFTGAAMVKGKFLI [114 - 127] INSERTION
# VP3 B-3: AYT [128 - 130] ABT CHAIN B
# VP3 I1-2: P [131] INSERTION
# VP3 B-4: PGAGK [132 - 136] ABT CHAIN B
# VP3 I123-3: P [137] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: T [138] INSERTION
# VP3 B-5: TRDQAM [139 - 144] ABT CHAIN B
# VP3 M-6: QATYAIWDLGLNSSFVFTAPFISPTH [145 - 170] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRQTSYTSATIAS (RES 180-181 'TI' MISSING FROM PDB) [171 - 183] INSERTION
# VP3 M-7: VD [184 - 185] MP2 NON-HELIX
# VP3 I123-5: GWVTVW [186 - 191] INSERTION
# VP3 M-8: Q [192] MP2 NON-HELIX
# VP3 I123-6: LT [193 - 194] INSERTION
# VP3 M-9: P [195] MP2 NON-HELIX
# VP3 I23-3: LT [196 - 197] INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: Y [198] MP2 NON-HELIX
# VP3 I13-2: PSG [199 - 201] INSERTION
# VP3 I123-7: TPV [202 - 204] INSERTION
# VP3 M-12: NSDILTLVSAG [205 - 215] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: DDFTLRMPISPTKWV [216 - 230] ABT CHAIN A
# VP3 M-13: PQ [231 - 232] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: GSDN (RES 233-236 'GSDN' MISSING FROM PDB) [233 - 236] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_E3_F3_1of6-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_E3_F3_2of6-1TME_VP3_TRIMER.pdb \
--remove="AEIBFJDHL";
# VP3 I123-1: Q [82] INSERTION
# VP3 I123-2: M [94] INSERTION
# VP3 I123-3: P [137] INSERTION
# VP3 I123-4: T [138] INSERTION
# VP3 I123-5: GWVTVW [186 - 191] INSERTION
# VP3 I123-6: LT [193 - 194] INSERTION
# VP3 I123-7: TPV [202 - 204] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E3_F3_2of6-1TME_VP3_TRIMER.pdb \
--remove=",-81;,83-93;,95-136;,139-185;,192;,195-201;,205-;" \
--output=new/figs/fig14/figure_14_panels_E3_F3_6of6-INSERT_I123_OF_1TME_VP3_TRIMER.pdb;
# VP3 I13-1: GSLN [107 - 110] INSERTION
# VP3 I13-2: PSG [199 - 201] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E3_F3_2of6-1TME_VP3_TRIMER.pdb \
--remove=",-106;,111-198;,202-;" \
--output=new/figs/fig14/figure_14_panels_E3_F3_4of6-INSERT_I13_OF_1TME_VP3_TRIMER.pdb;
# VP3 I23-1: CMCN [89 - 92] INSERTION
# VP3 I23-2: YRQTSYTSATIAS (RES 180-181 'TI' MISSING FROM PDB) [171 - 183] INSERTION
# VP3 I23-3: LT [196 - 197] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E3_F3_2of6-1TME_VP3_TRIMER.pdb \
--remove=",-88;,93-170;,184-195;,198-;" \
--output=new/figs/fig14/figure_14_panels_E3_F3_5of6-INSERT_I23_OF_1TME_VP3_TRIMER.pdb;
# VP3 M-1: PNSNNKRY [57 - 64] MP2 NON-HELIX
# VP3 I3-2: VALSCS [83 - 88] INSERTION
# VP3 I3-3: INSERTION
# VP3 I3-4: VFTGAAMVKGKFLI [114 - 127] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E3_F3_2of6-1TME_VP3_TRIMER.pdb \
--remove=",-56;,65-82;,89-113;,128-;" \
--output=new/figs/fig14/figure_14_panels_E3_F3_3of6-INSERT_I3_OF_1TME_VP3_TRIMER.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: MEMYRVR [65 - 71] MP2 NON-HELIX
# VP3 I3-1: LS [72 - 73] INSERTION
# VP3 M-2: DKPHTDD [74 - 80] MP2 HELIX ONE
# VP3 I123-1: INSERTION
# VP3 I3-2: PILCLSLS [81 - 88] INSERTION
# VP3 I23-1: PASD [89 - 92] INSERTION
# VP3 M-3: PR [93 - 94] MP2 HELIX ONE
# VP3 I123-2: INSERTION
# VP3 M-4: LSHT [95 - 98] MP2 HELIX TWO
# VP3 M-5: MLGEILNYYTH [99 - 109] MP2 NON-HELIX
# VP3 I3-3: WA [110 - 111] INSERTION
# VP3 I13-1: GSL [112 - 114] INSERTION
# VP3 B-1: ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: KFTF [115 - 118] ABT CHAIN B
# VP3 I3-4: LFCGSMMATGKLLV [119 - 132] INSERTION
# VP3 B-3: SYA [133 - 135] ABT CHAIN B
# VP3 I1-2: P [136] INSERTION
# VP3 B-4: PGAD [137 - 140] ABT CHAIN B
# VP3 I123-3: P [141] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: PK [142 - 143] INSERTION
# VP3 B-5: KRKEAML [144 - 150] ABT CHAIN B
# VP3 M-6: GTHVIWDIGLQSSCTMVVPWISNTT [151 - 175] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRQTI [176 - 180] INSERTION
# VP3 M-7: DDSFTE [181 - 186] MP2 NON-HELIX
# VP3 I123-5: GGYISVFY [187 - 194] INSERTION
# VP3 M-8: Q [195] MP2 NON-HELIX
# VP3 I123-6: T [196] INSERTION
# VP3 M-9: R [197] MP2 NON-HELIX
# VP3 I23-3: INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: IVV [198 - 200] MP2 NON-HELIX
# VP3 I13-2: PLS [201 - 203] INSERTION
# VP3 I123-7: TPR [204 - 206] INSERTION
# VP3 M-12: EMDILGFVSAC [207 - 217] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: NDFSVRLLR [218 - 226] ABT CHAIN A
# VP3 M-13: DTT [227 - 229] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: HIEQKALA (RES 236-237 'LA' MISSING) [230 - 237] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_E4_F4_1of6-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1HXS_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_E4_F4_2of6-1HXS_VP3_TRIMER.pdb \
--remove="AEIBFJDHL";
# VP3 I123-1: INSERTION
# VP3 I123-2: INSERTION
# VP3 I123-3: P [141] INSERTION
# VP3 I123-4: PK [142 - 143] INSERTION
# VP3 I123-5: GGYISVFY [187 - 194] INSERTION
# VP3 I123-6: T [196] INSERTION
# VP3 I123-7: TPR [204 - 206] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E4_F4_2of6-1HXS_VP3_TRIMER.pdb \
--remove=",-140;,144-186;,195;,197-203;,207-;" \
--output=new/figs/fig14/figure_14_panels_E4_F4_6of6-INSERT_I123_OF_1HXS_VP3_TRIMER.pdb;
# VP3 I13-1: GSL [112 - 114] INSERTION
# VP3 I13-2: PLS [201 - 203] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E4_F4_2of6-1HXS_VP3_TRIMER.pdb \
--remove=",-111;,115-200;,204-;" \
--output=new/figs/fig14/figure_14_panels_E4_F4_4of6-INSERT_I13_OF_1HXS_VP3_TRIMER.pdb;
# VP3 I23-1: PASD [89 - 92] INSERTION
# VP3 I23-2: YRQTI [176 - 180] INSERTION
# VP3 I23-3: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E4_F4_2of6-1HXS_VP3_TRIMER.pdb \
--remove=",-88;,93-175;,181-;" \
--output=new/figs/fig14/figure_14_panels_E4_F4_5of6-INSERT_I23_OF_1HXS_VP3_TRIMER.pdb;
# VP3 I3-1: LS [72 - 73] INSERTION
# VP3 I3-2: PILCLSLS [81 - 88] INSERTION
# VP3 I3-3: WA [110 - 111] INSERTION
# VP3 I3-4: LFCGSMMATGKLLV [119 - 132] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E4_F4_2of6-1HXS_VP3_TRIMER.pdb \
--remove=",-71;,74-80;,89-109;,112-118;,133-;" \
--output=new/figs/fig14/figure_14_panels_E4_F4_3of6-INSERT_I3_OF_1HXS_VP3_TRIMER.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP3 M-1: MERLRVD [64 - 70] MP2 NON-HELIX
# VP3 I3-1: ISVQA [71 - 75] INSERTION
# VP3 M-2: DLD [76 - 78] MP2 HELIX ONE
# VP3 I123-1: Q [79] INSERTION
# VP3 I3-2: LLFNIPLD [80 - 87] INSERTION
# VP3 I23-1: IQLD [88 - 91] INSERTION
# VP3 M-3: GPLRN [92 - 96] MP2 HELIX ONE
# VP3 I123-2: T [97] INSERTION
# VP3 M-4: LVGNISR [98 - 104] MP2 HELIX TWO
# VP3 M-5: YYTH [105 - 108] MP2 NON-HELIX
# VP3 I3-3: WS [109 - 110] INSERTION
# VP3 I13-1: GSLE [111 - 114] INSERTION
# VP3 B-1: ABT CHAIN B
# VP3 I1-1: INSERTION
# VP3 B-2: MTF [115 - 117] ABT CHAIN B
# VP3 I3-4: MFCGSFMATGKLILC [118 - 132] INSERTION
# VP3 B-3: YT [133 - 134] ABT CHAIN B
# VP3 I1-2: P [135] INSERTION
# VP3 B-4: PGGSC [136 - 140] ABT CHAIN B
# VP3 I123-3: P [141] INSERTION
# VP3 I1-3: INSERTION
# VP3 I123-4: T [142] INSERTION
# VP3 B-5: TRETAML [143 - 149] ABT CHAIN B
# VP3 M-6: GTHIVWDFGLQSSITLIIPWISGSH [150 - 174] MP2 NON-HELIX
# VP3 I2-1: INSERTION
# VP3 I23-2: YRMFNS [175 - 180] INSERTION
# VP3 M-7: DAKST [181 - 185] MP2 NON-HELIX
# VP3 I123-5: NANVGYVTCFM [186 - 196] INSERTION
# VP3 M-8: Q [197] MP2 NON-HELIX
# VP3 I123-6: T [198] INSERTION
# VP3 M-9: N [199] MP2 NON-HELIX
# VP3 I23-3: INSERTION
# VP3 I2-2: INSERTION
# VP3 M-10: MP2 NON-HELIX
# VP3 I2-3: INSERTION
# VP3 M-11: LIV [200 - 202] MP2 NON-HELIX
# VP3 I13-2: P [203] INSERTION
# VP3 I123-7: INSERTION
# VP3 M-12: SESSDTCSLIGFIAAK [204 - 219] MP2 NON-HELIX
# VP3 A-1: ABT CHAIN A
# VP3 I1-4: INSERTION
# VP3 A-2: DDFSLRLMR [220 - 228] ABT CHAIN A
# VP3 M-13: DSPD [229 - 232] MP2 NON-HELIX
# VP3 I2-4: INSERTION
# VP3 M-14: IGQSNHLHGAEAAYQ [233 - 247] MP2 NON-HELIX
# VP3 A-3: ABT CHAIN A
# VP3 I2-5: INSERTION
# VP3 A-4: ABT CHAIN A
# VP3 I1-5: INSERTION
# VP3 A-5: ABT CHAIN A
# VP3 M-15: MP2 NON-HELIX
# VP3 I2-6: INSERTION
# VP3 M-16: MP2 NON-HELIX
# VP3 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_E5_F5_1of6-ICOSAHEDRON.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_E5_F5_2of6-4WM7_VP3_TRIMER.pdb \
--remove="AEIBFJDHL";
# VP3 I123-1: Q [79] INSERTION
# VP3 I123-2: T [97] INSERTION
# VP3 I123-3: P [141] INSERTION
# VP3 I123-4: T [142] INSERTION
# VP3 I123-5: NANVGYVTCFM [186 - 196] INSERTION
# VP3 I123-6: T [198] INSERTION
# VP3 I123-7: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E5_F5_2of6-4WM7_VP3_TRIMER.pdb \
--remove=",-78;,80-96;,98-140;,143-185;,197;,199-;" \
--output=new/figs/fig14/figure_14_panels_E5_F5_6of6-INSERT_I123_OF_4WM7_VP3_TRIMER.pdb;
# VP3 I13-1: GSLE [111 - 114] INSERTION
# VP3 I13-2: P [203] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E5_F5_2of6-4WM7_VP3_TRIMER.pdb \
--remove=",-110;,115-202;,204-;" \
--output=new/figs/fig14/figure_14_panels_E5_F5_4of6-INSERT_I13_OF_4WM7_VP3_TRIMER.pdb;
# VP3 I23-1: IQLD [88 - 91] INSERTION
# VP3 I23-3: INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E5_F5_2of6-4WM7_VP3_TRIMER.pdb \
--remove=",-87;,92-;" \
--output=new/figs/fig14/figure_14_panels_E5_F5_5of6-INSERT_I23_OF_4WM7_VP3_TRIMER.pdb;
# VP3 I3-1: ISVQA [71 - 75] INSERTION
# VP3 I3-2: LLFNIPLD [80 - 87] INSERTION
# VP3 I3-3: WS [109 - 110] INSERTION
# VP3 I3-4: MFCGSFMATGKLILC [118 - 132] INSERTION
stripPDB --input=new/figs/fig14/figure_14_panels_E5_F5_2of6-4WM7_VP3_TRIMER.pdb \
--remove=",-70;,76-79;,88-108;,111-117;,133-;" \
--output=new/figs/fig14/figure_14_panels_E5_F5_3of6-INSERT_I3_OF_4WM7_VP3_TRIMER.pdb;
# Combine related substructures of VP1, VP2, and VP3 to make Figure 14 VP123 substructures.
# FMDV (1BBT) VP123 TRIMER
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_G1_H1_1of12-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1BBT_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_G1_H1_2of12-1BBT_VP123_TRIMER.pdb \
--remove="DHL";
cp new/figs/fig14/figure_14_panels_A1_B1_3of5-INSERT_I1_OF_1BBT_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_3of12-INSERT_I1_OF_1BBT_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C1_D1_3of5-INSERT_I2_OF_1BBT_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_4of12-INSERT_I2_OF_1BBT_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E1_F1_3of6-INSERT_I3_OF_1BBT_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_5of12-INSERT_I3_OF_1BBT_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A1_B1_4of5-INSERT_I13_OF_1BBT_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_6of12-INSERT_I13_OF_1BBT_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E1_F1_4of6-INSERT_I13_OF_1BBT_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_7of12-INSERT_I13_OF_1BBT_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C1_D1_4of5-INSERT_I23_OF_1BBT_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_8of12-INSERT_I23_OF_1BBT_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E1_F1_5of6-INSERT_I23_OF_1BBT_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_9of12-INSERT_I23_OF_1BBT_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A1_B1_5of5-INSERT_I123_OF_1BBT_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_10of12-INSERT_I123_OF_1BBT_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C1_D1_5of5-INSERT_I123_OF_1BBT_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_11of12-INSERT_I123_OF_1BBT_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E1_F1_6of6-INSERT_I123_OF_1BBT_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G1_H1_12of12-INSERT_I123_OF_1BBT_VP3_TRIMER.pdb;
# TMEV-2 (1TMF) VP123 TRIMER
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_G2_H2_1of12-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TMF_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_G2_H2_2of12-1TMF_VP123_TRIMER.pdb \
--remove="DHL";
cp new/figs/fig14/figure_14_panels_A2_B2_3of5-INSERT_I1_OF_1TMF_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_3of12-INSERT_I1_OF_1TMF_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C2_D2_3of5-INSERT_I2_OF_1TMF_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_4of12-INSERT_I2_OF_1TMF_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E2_F2_3of6-INSERT_I3_OF_1TMF_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_5of12-INSERT_I3_OF_1TMF_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A2_B2_4of5-INSERT_I13_OF_1TMF_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_6of12-INSERT_I13_OF_1TMF_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E2_F2_4of6-INSERT_I13_OF_1TMF_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_7of12-INSERT_I13_OF_1TMF_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C2_D2_4of5-INSERT_I23_OF_1TMF_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_8of12-INSERT_I23_OF_1TMF_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E2_F2_5of6-INSERT_I23_OF_1TMF_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_9of12-INSERT_I23_OF_1TMF_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A2_B2_5of5-INSERT_I123_OF_1TMF_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_10of12-INSERT_I123_OF_1TMF_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C2_D2_5of5-INSERT_I123_OF_1TMF_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_11of12-INSERT_I123_OF_1TMF_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E2_F2_6of6-INSERT_I123_OF_1TMF_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G2_H2_12of12-INSERT_I123_OF_1TMF_VP3_TRIMER.pdb;
# TMEV-1 (1TME) VP123 TRIMER
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_G3_H3_1of12-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1TME_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_G3_H3_2of12-1TME_VP123_TRIMER.pdb \
--remove="DHL";
cp new/figs/fig14/figure_14_panels_A3_B3_3of5-INSERT_I1_OF_1TME_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_3of12-INSERT_I1_OF_1TME_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C3_D3_3of5-INSERT_I2_OF_1TME_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_4of12-INSERT_I2_OF_1TME_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E3_F3_3of6-INSERT_I3_OF_1TME_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_5of12-INSERT_I3_OF_1TME_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A3_B3_4of5-INSERT_I13_OF_1TME_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_6of12-INSERT_I13_OF_1TME_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E3_F3_4of6-INSERT_I13_OF_1TME_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_7of12-INSERT_I13_OF_1TME_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C3_D3_4of5-INSERT_I23_OF_1TME_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_8of12-INSERT_I23_OF_1TME_VP2_TRIMER.pdb
cp new/figs/fig14/figure_14_panels_E3_F3_5of6-INSERT_I23_OF_1TME_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_9of12-INSERT_I23_OF_1TME_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A3_B3_5of5-INSERT_I123_OF_1TME_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_10of12-INSERT_I123_OF_1TME_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C3_D3_5of5-INSERT_I123_OF_1TME_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_11of12-INSERT_I123_OF_1TME_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E3_F3_6of6-INSERT_I123_OF_1TME_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G3_H3_12of12-INSERT_I123_OF_1TME_VP3_TRIMER.pdb;
# POLIO-Mahoney (1HXS) VP123 TRIMER
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_G4_H4_1of12-ICOSAHEDRON.pdb;
stripPDB --input=new/super/1HXS_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_G4_H4_2of12-1HXS_VP123_TRIMER.pdb \
--remove="DHL";
cp new/figs/fig14/figure_14_panels_A4_B4_3of5-INSERT_I1_OF_1HXS_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_3of12-INSERT_I1_OF_1HXS_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C4_D4_3of5-INSERT_I2_OF_1HXS_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_4of12-INSERT_I2_OF_1HXS_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E4_F4_3of6-INSERT_I3_OF_1HXS_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_5of12-INSERT_I3_OF_1HXS_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A4_B4_4of5-INSERT_I13_OF_1HXS_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_6of12-INSERT_I13_OF_1HXS_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E4_F4_4of6-INSERT_I13_OF_1HXS_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_7of12-INSERT_I13_OF_1HXS_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C4_D4_4of5-INSERT_I23_OF_1HXS_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_8of12-INSERT_I23_OF_1HXS_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E4_F4_5of6-INSERT_I23_OF_1HXS_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_9of12-INSERT_I23_OF_1HXS_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A4_B4_5of5-INSERT_I123_OF_1HXS_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_10of12-INSERT_I123_OF_1HXS_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C4_D4_5of5-INSERT_I123_OF_1HXS_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_11of12-INSERT_I123_OF_1HXS_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E4_F4_6of6-INSERT_I123_OF_1HXS_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G4_H4_12of12-INSERT_I123_OF_1HXS_VP3_TRIMER.pdb;
# EV-D68 (4WM7) VP123 TRIMER
cp new/tmpicosframe.pdb new/figs/fig14/figure_14_panels_G5_H5_1of12-ICOSAHEDRON.pdb;
stripPDB --input=new/super/4WM7_tile_1.pdb \
--output=new/figs/fig14/figure_14_panels_G5_H5_2of12-4WM7_VP123_TRIMER.pdb \
--remove="DHL";
cp new/figs/fig14/figure_14_panels_A5_B5_3of5-INSERT_I1_OF_4WM7_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_3of12-INSERT_I1_OF_4WM7_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C5_D5_3of5-INSERT_I2_OF_4WM7_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_4of12-INSERT_I2_OF_4WM7_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E5_F5_3of6-INSERT_I3_OF_4WM7_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_5of12-INSERT_I3_OF_4WM7_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A5_B5_4of5-INSERT_I13_OF_4WM7_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_6of12-INSERT_I13_OF_4WM7_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E5_F5_4of6-INSERT_I13_OF_4WM7_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_7of12-INSERT_I13_OF_4WM7_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C5_D5_4of5-INSERT_I23_OF_4WM7_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_8of12-INSERT_I23_OF_4WM7_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E5_F5_5of6-INSERT_I23_OF_4WM7_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_9of12-INSERT_I23_OF_4WM7_VP3_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_A5_B5_5of5-INSERT_I123_OF_4WM7_VP1_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_10of12-INSERT_I123_OF_4WM7_VP1_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_C5_D5_5of5-INSERT_I123_OF_4WM7_VP2_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_11of12-INSERT_I123_OF_4WM7_VP2_TRIMER.pdb;
cp new/figs/fig14/figure_14_panels_E5_F5_6of6-INSERT_I123_OF_4WM7_VP3_TRIMER.pdb \
new/figs/fig14/figure_14_panels_G5_H5_12of12-INSERT_I123_OF_4WM7_VP3_TRIMER.pdb;
# Figure 15. Ion channel-like structure in the center of myelin P2 trimers and picornavirus VP1 trimers.
#
# Atoms are stripped from Myelin P2 trimers to make trimer substructures
# showing Myelin P2 Channel Sections and Myelin P2 Helix Sections.
#
# Atoms are stripped from Picornavirus trimers to make VP1 trimer substructures showing
# residues corresponding to Myelin P2 Channel Sections and Myelin P2 Helix Sections.
#
# "HELIX" Sections: M-2, M-3, M-4
# "CHANNEL" Sections: M-5, M-6, M-7, M-8, M-9, M-10, M-14
#
# The following is useful for panels in rows B - F:
#
# VP1234 chain identifier ordinal: 1 2 3 4
# First VP1234 chain identifiers: A B C D
# Second VP1234 chain identifiers: E F G H
# Third VP1234 chain identifiers: I J K L
# Strip atoms to make Figure 15 substructures of a myelin P2 trimer.
#===================================================================================================
# 2WUT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# M-1: GMSNKFLGTWKLVS [(-1) - 12] MP2 NON-HELIX
# M-2: SENFDDYM [13 - 20] MP2 HELIX ONE
# M-3: KALG [21 - 24] MP2 HELIX ONE
# M-4: VGLATRKLGNLAK [25 - 37] MP2 HELIX TWO
# M-5: PTVIISKKGDIITIRTEST [38 - 56] MP2 NON-HELIX
# M-6: FKNTEISFKLG [57 - 67] MP2 NON-HELIX
# M-7: QEFEETT [68 - 74] MP2 NON-HELIX
# M-8: ADN [75 - 77] MP2 NON-HELIX
# M-9: RKT [78 - 80] MP2 NON-HELIX
# M-10: KS [81 - 82] MP2 NON-HELIX
# M-11: IV [83 - 84] MP2 NON-HELIX
# M-12: TLQRGSLNQVQR [85 - 96] MP2 NON-HELIX
# M-13: WDGKETTI [97 - 104] MP2 NON-HELIX
# M-14: KRKLVNGKMVAECKMK [105 - 120] MP2 NON-HELIX
# M-15: GVVCT [121 - 125] MP2 NON-HELIX
# M-16: RIYEKV [126 - 131] MP2 NON-HELIX
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_A1_A3_1of2-ICOSAHEDRON.pdb;
# M-5: PTVIISKKGDIITIRTEST [38 - 56] MP2 NON-HELIX
# M-6: FKNTEISFKLG [57 - 67] MP2 NON-HELIX
# M-7: QEFEETT [68 - 74] MP2 NON-HELIX
# M-8: ADN [75 - 77] MP2 NON-HELIX
# M-9: RKT [78 - 80] MP2 NON-HELIX
# M-10: KS [81 - 82] MP2 NON-HELIX
# M-13: WDGKETTI [97 - 104] MP2 NON-HELIX
# M-14: KRKLVNGKMVAECKMK [105 - 120] MP2 NON-HELIX
stripPDB --input=new/super/2WUT_TRIMER_table5.pdb --remove="\
A,-37;A,83-96;A,121-;\
B,-37;B,83-96;B,121-;\
C,-37;C,83-96;C,121-;"\
--output=new/figs/fig15/figure_15_panels_A1_A3_2of2-MP2_2WUT_CHANNEL_TRIMER.pdb;
# M-2: SENFDDYM [13 - 20] MP2 HELIX ONE
# M-3: KALG [21 - 24] MP2 HELIX ONE
# M-4: VGLATRKLGNLAK [25 - 37] MP2 HELIX TWO
# M-5: PTVIISKKGDIITIRTEST [38 - 56] MP2 NON-HELIX
# M-6: FKNTEISFKLG [57 - 67] MP2 NON-HELIX
# M-7: QEFEETT [68 - 74] MP2 NON-HELIX
# M-8: ADN [75 - 77] MP2 NON-HELIX
# M-9: RKT [78 - 80] MP2 NON-HELIX
# M-10: KS [81 - 82] MP2 NON-HELIX
# M-13: WDGKETTI [97 - 104] MP2 NON-HELIX
# M-14: KRKLVNGKMVAECKMK [105 - 120] MP2 NON-HELIX
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_A2_A4_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig15/figure_15_panels_A1_A3_2of2-MP2_2WUT_CHANNEL_TRIMER.pdb \
new/figs/fig15/figure_15_panels_A2_A4_2of3-MP2_2WUT_CHANNEL_TRIMER.pdb;
stripPDB --input=new/super/2WUT_TRIMER_table5.pdb --remove="\
A,-12;A,38-;\
B,-12;B,38-;\
C,-12;C,38-;"\
--output=new/figs/fig15/figure_15_panels_A2_A4_3of3-MP2_2WUT_HELIX_TRIMER.pdb;
# Strip atoms to make Figure 15 VP1 substructures.
#===================================================================================================
# 1BBT.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: TTSAGESADPVTTTVENYGG [1 - 20] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: ETQI [21 - 24] MP2 HELIX ONE
# VP1 I123-1: Q [25] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: RR [26 - 27] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHTDVSFIMDR [28 - 38] MP2 HELIX TWO
# VP1 M-5: FVKVTPQNQI [39 - 48] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: N [49] INSERTION
# VP1 B-1: IL [50 - 51] ABT CHAIN B
# VP1 I1-1: INSERTION
# VP1 B-2: ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: ABT CHAIN B
# VP1 I1-2: D [52] INSERTION
# VP1 B-4: LM [53 - 54] ABT CHAIN B
# VP1 I123-3: QVPSHT [55 - 60] INSERTION
# VP1 I1-3: LV [61 - 62] INSERTION
# VP1 I123-4: GG [63 - 64] INSERTION
# VP1 B-5: LLRAS [65 - 69] ABT CHAIN B
# VP1 M-6: TYYFSDLEIAVKH [70 - 82] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: EG [83 - 84] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: D [85] MP2 NON-HELIX
# VP1 I123-6: LT [86 - 87] INSERTION
# VP1 M-9: MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WV [88 - 89] MP2 NON-HELIX
# VP1 I13-2: PNGA [90 - 93] INSERTION
# VP1 I123-7: PE [94 - 95] INSERTION
# VP1 M-12: KALDNTTNPTAYHKAPLTR [96 - 114] MP2 NON-HELIX
# VP1 A-1: LALPYTAPHRVLATVYNGECRYSRNAV (PDB MISSING 'RYSRNAV') [115 - 141] ABT CHAIN A
# VP1 I1-4: PNLRGDLQVLAQ (PDB MISSING 'PNLRGDLQVLAQ') [142 - 153] INSERTION
# VP1 A-2: KVARTLPTSFNY (RES 154 - 156 'KVA' MISSING FROM PDB) [154 - 165] ABT CHAIN A
# VP1 M-13: GAIKAT [166 - 171] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: RVTELLYRMKRAETYCPRPLLA [172 - 193] MP2 NON-HELIX
# VP1 A-3: I [194] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: H [195] ABT CHAIN A
# VP1 M-15: PTEA [196 - 199] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RHKQKIVAPVK (PDB MISSING 'VK') [200 - 210] MP2 NON-HELIX
# VP1 A-6: QTL (PDB MISSING 'QTL') [211 - 213] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_B1_B3_1of2-ICOSAHEDRON.pdb;
# VP1 M-5: FVKVTPQNQI [39 - 48] MP2 NON-HELIX
# VP1 M-6: TYYFSDLEIAVKH [70 - 82] MP2 NON-HELIX
# VP1 M-7: EG [83 - 84] MP2 NON-HELIX
# VP1 M-8: D [85] MP2 NON-HELIX
# VP1 M-9: MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-13: GAIKAT [166 - 171] MP2 NON-HELIX
# VP1 M-14: RVTELLYRMKRAETYCPRPLLA [172 - 193] MP2 NON-HELIX
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,-38;A,49-69;A,86-165;A,194-;B;C;D;\
E,-38;E,49-69;E,86-165;E,194-;F;G;H;\
I,-38;I,49-69;I,86-165;I,194-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_B1_B3_2of2-MP2_CHANNEL_OF_1BBT_VP1_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_B2_B4_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig15/figure_15_panels_B1_B3_2of2-MP2_CHANNEL_OF_1BBT_VP1_TRIMER.pdb \
new/figs/fig15/figure_15_panels_B2_B4_2of3-MP2_CHANNEL_OF_1BBT_VP1_TRIMER.pdb;
# VP1 M-2: ETQI [21 - 24] MP2 HELIX ONE
# VP1 M-3: RR [26 - 27] MP2 HELIX ONE
# VP1 M-4: QHTDVSFIMDR [28 - 38] MP2 HELIX TWO
stripPDB --input=new/super/1BBT_tile_1.pdb --remove="\
A,-20;A,25;A,39-;B;C;D;\
E,-20;E,25;E,39-;F;G;H;\
I,-20;I,25;I,39-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_B2_B4_3of3-MP2_HELIX_OF_1BBT_VP1_TRIMER.pdb;
#===================================================================================================
# 1TMF.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GVDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FNY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDYRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSSSGPQKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGVN [99 - 102] INSERTION
# VP1 I123-4: GAN [103 - 105] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [106 - 120] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 I123-6: ASVL [142 - 145] INSERTION
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [147 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGA [149 - 152] INSERTION
# VP1 I123-7: PAD [153 - 155] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [156 - 176] MP2 NON-HELIX
# VP1 A-1: LVGAGNSQVSFVVPYNSPLSVLPAAWFNGWS [177 - 207] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [209 - 220] INSERTION
# VP1 A-2: ADFGRLWI [221 - 228] ABT CHAIN A
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [251 - 256] MP2 NON-HELIX
# VP1 I2-6: PT [257 - 258] INSERTION
# VP1 M-16: PTTTKINADNPVPILELE [259 - 276] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_C1_C3_1of2-ICOSAHEDRON.pdb;
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-39;A,56-120;A,142-145;A,147-228;A,251-;B;C;D;\
E,-39;E,56-120;E,142-145;E,147-228;E,251-;F;G;H;\
I,-39;I,56-120;I,142-145;I,147-228;I,251-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_C1_C3_2of2-MP2_CHANNEL_OF_1TMF_VP1_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_C2_C4_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig15/figure_15_panels_C1_C3_2of2-MP2_CHANNEL_OF_1TMF_VP1_TRIMER.pdb \
new/figs/fig15/figure_15_panels_C2_C4_2of3-MP2_CHANNEL_OF_1TMF_VP1_TRIMER.pdb;
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-10;A,22;A,27-28;A,40-;B;C;D;\
E,-10;E,22;E,27-28;E,40-;F;G;H;\
I,-10;I,22;I,27-28;I,40-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_C2_C4_3of3-MP2_HELIX_OF_1TMF_VP1_TRIMER.pdb;
#===================================================================================================
# 1TME.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: GSDNAEKGKV [1 - 10] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 I123-1: E [22] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 I123-2: PE [27 - 28] INSERTION
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
# VP1 M-5: AVPIGMLRPGQNIEST [40 - 55] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FVY [56 - 58] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: QENDLRLN [59 - 66] INSERTION
# VP1 B-2: CLLLTPL [67 - 73] ABT CHAIN B
# VP1 I3-4: P [74] INSERTION
# VP1 B-3: SFC [75 - 77] ABT CHAIN B
# VP1 I1-2: PDSTSGPVKTKA [78 - 89] INSERTION
# VP1 B-4: PVQWRW [90 - 95] ABT CHAIN B
# VP1 I123-3: VRS [96 - 98] INSERTION
# VP1 I1-3: GGT [99 - 101] INSERTION
# VP1 I123-4: TN [102 - 103] INSERTION
# VP1 B-5: FPLMTKQDYAFLCFS [104 - 118] ABT CHAIN B
# VP1 M-6: PFTYYKCDLEVTVSAL [119 - 134] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: GTDT [135 - 138] MP2 NON-HELIX
# VP1 I123-5: INSERTION
# VP1 M-8: V [139] MP2 NON-HELIX
# VP1 I123-6: ASVL [140 - 143] INSERTION
# VP1 M-9: R [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: WA [145 - 146] MP2 NON-HELIX
# VP1 I13-2: PTGA [147 - 150] INSERTION
# VP1 I123-7: PAD [151 - 153] INSERTION
# VP1 M-12: VTDQLIGYTPSLGETRNPHMW [154 - 174] MP2 NON-HELIX
# VP1 A-1: LVGAGNTQISFVVPYNSPLSVLPAAWFNGWS [175 - 205] ABT CHAIN A
# VP1 I1-4: DFGNTKDFGVAPN [206 - 218] INSERTION
# VP1 A-2: ADFGRLWI [219 - 226] ABT CHAIN A
# VP1 M-13: QGNTSA [227 - 232] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: SVRIRYKKMKVFCPRP [233 - 248] MP2 NON-HELIX
# VP1 A-3: ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: ABT CHAIN A
# VP1 I1-5: INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: TLFFPW [249 - 254] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: PVSTRSKINADNPVPILELE [255 - 274] MP2 NON-HELIX
# VP1 A-6: ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_D1_D3_1of2-ICOSAHEDRON.pdb;
# VP1 M-5: AVPIGMLRPGQNIEST [40 - 55] MP2 NON-HELIX
# VP1 M-6: PFTYYKCDLEVTVSAL [119 - 134] MP2 NON-HELIX
# VP1 M-7: GTDT [135 - 138] MP2 NON-HELIX
# VP1 M-8: V [139] MP2 NON-HELIX
# VP1 M-9: R [144] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-13: QGNTSA [227 - 232] MP2 NON-HELIX
# VP1 M-14: SVRIRYKKMKVFCPRP [233 - 248] MP2 NON-HELIX
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,-39;A,56-118;A,140-143;A,145-226;A,249-;B;C;D;\
E,-39;E,56-118;E,140-143;E,145-226;E,249-;F;G;H;\
I,-39;I,56-118;I,140-143;I,145-226;I,249-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_D1_D3_2of2-MP2_CHANNEL_OF_1TME_VP1_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_D2_D4_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig15/figure_15_panels_D1_D3_2of2-MP2_CHANNEL_OF_1TME_VP1_TRIMER.pdb \
new/figs/fig15/figure_15_panels_D2_D4_2of3-MP2_CHANNEL_OF_1TME_VP1_TRIMER.pdb;
# VP1 M-2: SNDDASVDFVA [11 - 21] MP2 HELIX ONE
# VP1 M-3: PVKL [23 - 26] MP2 HELIX ONE
# VP1 M-4: NQTRVAFFYDR [29 - 39] MP2 HELIX TWO
stripPDB --input=new/super/1TME_tile_1.pdb --remove="\
A,-10;A,22;A,27-28;A,40-;B;C;D;\
E,-10;E,22;E,27-28;E,40-;F;G;H;\
I,-10;I,22;I,27-28;I,40-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_D2_D4_3of3-MP2_HELIX_OF_1TME_VP1_TRIMER.pdb;
#===================================================================================================
# 4WM7.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PSLNAVETGATSN [24 - 36] MP2 NON-HELIX
# VP1 I3-1: INSERTION
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 I123-1: Q [44] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: FEY [74 - 76] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: KN [77 - 78] INSERTION
# VP1 B-2: HAS (RES 81 'S' MISSING FROM PDB) [79 - 81] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: SSA (RES 82-84 'SSA' MISSING FROM PDB) [82 - 84] ABT CHAIN B
# VP1 I1-2: GTHK (RES 81 'GT' MISSING FROM PDB) [85 - 88] INSERTION
# VP1 B-4: NFFKW [89 - 93] ABT CHAIN B
# VP1 I123-3: T [94] INSERTION
# VP1 I1-3: INT [95 - 97] INSERTION
# VP1 I123-4: K [98] INSERTION
# VP1 B-5: SFVQLRRKLEL [99 - 109] ABT CHAIN B
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 I123-5: YMGL [135 - 138] INSERTION
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 I123-6: LTL [141 - 143] INSERTION
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: AMFV [145 - 148] MP2 NON-HELIX
# VP1 I13-2: PTGAL [149 - 153] INSERTION
# VP1 I123-7: TP [154 - 155] INSERTION
# VP1 M-12: KEQDSFHWQSGSNASVFFKISDPPARM [156 - 182] MP2 NON-HELIX
# VP1 A-1: TIPFMCINSAYSVFYDGFA [183 - 201] ABT CHAIN A
# VP1 I1-4: GFEKNGLYGINPA [202 - 214] INSERTION
# VP1 A-2: DTIGNLCVRI [215 - 224] ABT CHAIN A
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 I2-4: G [232] INSERTION
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
# VP1 A-3: PYMSIA [258 - 263] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: NANY [264 - 267] ABT CHAIN A
# VP1 I1-5: KGRD [268 - 271] INSERTION
# VP1 A-5: TA [272 - 273] ABT CHAIN A
# VP1 M-15: PNTLNAIIGN [274 - 283] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: RASVTTM [284 - 290] MP2 NON-HELIX
# VP1 A-6: PHNIVTT (RES 297 'T' MISSING FROM PDB) [291 - 297] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_E1_E3_1of2-ICOSAHEDRON.pdb;
# VP1 M-5: ETLVENFLGRAALVSKKS [56 - 73] MP2 NON-HELIX
# VP1 M-6: FTYLRFDAEITILTTVAV [110 - 127] MP2 NON-HELIX
# VP1 M-7: NGNNDST (RES 129 - 134 'GNNDST' MISSING FROM PDB) [128 - 134] MP2 NON-HELIX
# VP1 M-8: PD [139 - 140] MP2 NON-HELIX
# VP1 M-9: Q [144] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-13: VNEHQPV [225 - 231] MP2 NON-HELIX
# VP1 M-14: FTVTVRVYMKPKHIKAWAPRPPRTM [233 - 257] MP2 NON-HELIX
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-55;A,74-109;A,135-138;A,141-143;A,145-224;A,232;A,258-;B;C;D;\
E,-55;E,74-109;E,135-138;E,141-143;E,145-224;E,232;E,258-;F;G;H;\
I,-55;I,74-109;I,135-138;I,141-143;I,145-224;I,232;I,258-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_E1_E3_2of2-MP2_CHANNEL_OF_4WM7_VP1_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_E2_E4_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig15/figure_15_panels_E1_E3_2of2-MP2_CHANNEL_OF_4WM7_VP1_TRIMER.pdb \
new/figs/fig15/figure_15_panels_E2_E4_2of3-MP2_CHANNEL_OF_4WM7_VP1_TRIMER.pdb;
# VP1 M-2: TEPEEAI [37 - 43] MP2 HELIX ONE
# VP1 M-3: TRTV [45 - 48] MP2 HELIX ONE
# VP1 M-4: INQHGVS [49 - 55] MP2 HELIX TWO
stripPDB --input=new/super/4WM7_tile_1.pdb --remove="\
A,-36;A,44;A,56-;B;C;D;\
E,-36;E,44;E,56-;F;G;H;\
I,-36;I,44;I,56-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_E2_E4_3of3-MP2_HELIX_OF_4WM7_VP1_TRIMER.pdb;
#===================================================================================================
# 1HXS.pdb sequence alignment and functional characterization from:
#
# Weininger, A.; Weininger, S. (2016)
# "Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural
# Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction"
# http://www.weiningerworks.com/picornavirus_monograph.html
#
# --------------------------------------------------------------------------------------------------
# SECTION SEQUENCE RESIDUE # SECTION DESCRIPTOR
# ----------- --------------------------------------------------- ----------- -------------------
# VP1 M-1: PALTAVETGATNPLV [43 - 56] MP2 NON-HELIX
# VP1 I3-1: P [57] INSERTION
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 I123-1: Q [62] INSERTION
# VP1 I3-2: INSERTION
# VP1 I23-1: INSERTION
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
# VP1 I123-2: INSERTION
# VP1 M-4: QHRSRS [68 - 73] MP2 HELIX TWO
# VP1 M-5: ESSIESFFARGACVTIMT [74 - 91] MP2 NON-HELIX
# VP1 I3-3: INSERTION
# VP1 I13-1: V [92] INSERTION
# VP1 B-1: ABT CHAIN B
# VP1 I1-1: DN [93 - 94] INSERTION
# VP1 B-2: PAS [95 - 97] ABT CHAIN B
# VP1 I3-4: INSERTION
# VP1 B-3: TT [98 - 99] ABT CHAIN B
# VP1 I1-2: NKDK [100 - 103] INSERTION
# VP1 B-4: LFAVW [104 - 108] ABT CHAIN B
# VP1 I123-3: K [109] INSERTION
# VP1 I1-3: IT [110 - 111] INSERTION
# VP1 I123-4: YK [112 - 113] INSERTION
# VP1 B-5: DTVQLRRKLEF [114 - 124] ABT CHAIN B
# VP1 M-6: FTYSRFDMELTFVVTA [125 - 140] MP2 NON-HELIX
# VP1 I2-1: INSERTION
# VP1 I23-2: INSERTION
# VP1 M-7: NFTETNN [141 - 147] MP2 NON-HELIX
# VP1 I123-5: GHA [148 - 150] INSERTION
# VP1 M-8: LNQ [151 - 153] MP2 NON-HELIX
# VP1 I123-6: VY [154 - 155] INSERTION
# VP1 M-9: Q [156] MP2 NON-HELIX
# VP1 I23-3: INSERTION
# VP1 I2-2: INSERTION
# VP1 M-10: MP2 NON-HELIX
# VP1 I2-3: INSERTION
# VP1 M-11: IMYV [157 - 160] MP2 NON-HELIX
# VP1 I13-2: PPGA [161 - 164] INSERTION
# VP1 I123-7: PVPE [165 - 168] INSERTION
# VP1 M-12: KWDDYTWQTSSNPSIFYTYGTAPAR [169 - 193] MP2 NON-HELIX
# VP1 A-1: ISVPYVGISNAYSHFYDGFSKV [194 - 215] ABT CHAIN A
# VP1 I1-4: PLKDQSAALGDSLYGAASLN [216 - 235] INSERTION
# VP1 A-2: DFGILAVRV [236 - 244] ABT CHAIN A
# VP1 M-13: VNDHNPT [245 - 251] MP2 NON-HELIX
# VP1 I2-4: INSERTION
# VP1 M-14: KVTSKIRVYLKPKHIRVWCPRPPRAVA [252 - 278] MP2 NON-HELIX
# VP1 A-3: YYG [279 - 281] ABT CHAIN A
# VP1 I2-5: INSERTION
# VP1 A-4: PGVDY [282 - 286] ABT CHAIN A
# VP1 I1-5: KD [287 - 288] INSERTION
# VP1 A-5: ABT CHAIN A
# VP1 M-15: GTLTP [289 - 293] MP2 NON-HELIX
# VP1 I2-6: INSERTION
# VP1 M-16: LSTKD [294 - 298] MP2 NON-HELIX
# VP1 A-6: LTTY [299 - 302] ABT CHAIN A
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_F1_F3_1of2-ICOSAHEDRON.pdb;
# VP1 M-5: ESSIESFFARGACVTIMT [74 - 91] MP2 NON-HELIX
# VP1 M-6: FTYSRFDMELTFVVTA [125 - 140] MP2 NON-HELIX
# VP1 M-7: NFTETNN [141 - 147] MP2 NON-HELIX
# VP1 M-8: LNQ [151 - 153] MP2 NON-HELIX
# VP1 M-9: Q [156] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-13: VNDHNPT [245 - 251] MP2 NON-HELIX
# VP1 M-14: KVTSKIRVYLKPKHIRVWCPRPPRAVA [252 - 278] MP2 NON-HELIX
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-73;A,92-124;A,148-150;A,154-155;A,157-244;A,279-;B;C;D;\
E,-73;E,92-124;E,148-150;E,154-155;E,157-244;E,279-;F;G;H;\
I,-73;I,92-124;I,148-150;I,154-155;I,157-244;I,279-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_F1_F3_2of2-MP2_CHANNEL_OF_1HXS_VP1_TRIMER.pdb;
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_F2_F4_1of3-ICOSAHEDRON.pdb;
cp new/figs/fig15/figure_15_panels_F1_F3_2of2-MP2_CHANNEL_OF_1HXS_VP1_TRIMER.pdb \
new/figs/fig15/figure_15_panels_F2_F4_2of3-MP2_CHANNEL_OF_1HXS_VP1_TRIMER.pdb;
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
# VP1 M-4: QHRSRS [68 - 73] MP2 HELIX TWO
stripPDB --input=new/super/1HXS_tile_1.pdb --remove="\
A,-57;A,62;A,74-;B;C;D;\
E,-57;E,62;E,74-;F;G;H;\
I,-57;I,62;I,74-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_F2_F4_3of3-MP2_HELIX_OF_1HXS_VP1_TRIMER.pdb;
# Strip atoms to show the potassium channel (KCSA) from streptomyces lividans (1BL8.pdb).
# 1BV8.pdb POTASSIUM ION CHANNEL: 53 to P83 (ON 4 IDENTICAL CHAINS MAKING A TETRAMER)
stripPDB --input=orig/1BL8.pdb --remove=",-52;,84-119;,402-;" \
--output=new/figs/fig15/figure_15_panels_G1_G2-K_ION_CHANNEL_OF_1BL8_TETRAMER.pdb;
# Strip atoms to show the potassium-like channel in TMEV-2 (1TMF.pdb).
cp new/tmpicosframe.pdb new/figs/fig15/figure_15_panels_G3_G4_1of4-ICOSAHEDRON.pdb;
# VP1 M-5: AVPIGMLRPGQNMETT [40 - 55] MP2 NON-HELIX
# VP1 M-6: PFTYYKCDLEVTVSAL [121 - 136] MP2 NON-HELIX
# VP1 M-7: GTDT [137 - 140] MP2 NON-HELIX
# VP1 M-8: V [141] MP2 NON-HELIX
# VP1 M-9: R [146] MP2 NON-HELIX
# VP1 M-10: MP2 NON-HELIX
# VP1 M-13: QGNTSA [229 - 234] MP2 NON-HELIX
# VP1 M-14: SVRIRYKKMKVFCPRP [235 - 250] MP2 NON-HELIX
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-39;A,56-120;A,142-145;A,147-228;A,251-;B;C;D;\
E,-39;E,56-120;E,142-145;E,147-228;E,251-;F;G;H;\
I,-39;I,56-120;I,142-145;I,147-228;I,251-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_G3_G4_2of4-MP2_CHANNEL_OF_1TMF_VP1_TRIMER.pdb;
# VP1 M-2: SDTV [58 - 61] MP2 HELIX ONE
# VP1 M-3: TRHVV [63 - 67] MP2 HELIX ONE
stripPDB --input=new/super/1TMF_tile_1.pdb --remove="\
A,-57;A,62;A,68-;B;C;D;\
E,-57;E,62;E,68-;F;G;H;\
I,-57;I,62;I,68-;J;K;L;"\
--output=new/figs/fig15/figure_15_panels_G3_G4_3of4-MP2_HELIX_ONE_OF_1TMF_VP1_TRIMER.pdb;
# Create PDB file to contain potassium ion positioned in center of MP2_CHANNEL_OF_1TMF_VP1_TRIMER.
#
#
# ATOM 1 O ALA A 40 -1.074 33.149 93.993 1.00 0.00 O
# ATOM 2 CG2 VAL A 41 -2.478 35.475 95.724 1.00 0.00 C
# ATOM 3 O ALA E 40 -1.690 37.622 92.284 1.00 0.00 O
# ATOM 4 CG2 VAL E 41 0.358 38.905 94.414 1.00 0.00 C
# ATOM 5 O ALA I 40 2.765 35.883 92.947 1.00 0.00 O
# ATOM 6 CG2 VAL I 41 2.120 34.894 95.945 1.00 0.00 C
#
# (((-1.074 - 1.690 + 2.765)/3) + ((-2.478 + 0.358 + 2.120)/3))/2 = 0.0001666666
# (((33.149 + 37.622 + 35.883)/3) + ((35.475 + 38.905 + 34.894)/3))/2 = 35.987999999
# (((93.993 + 92.284 + 92.947)/3) + ((95.724 + 94.414 + 95.945)/3))/2 = 94.217833333
printf 'REMARK 250 MODELED K ION FOR VIEWING WITH: \n' > new/tmpk;
printf 'REMARK 250 figure_15_panels_G3_G4_2of4-MP2_CHANNEL_OF_1TMF_VP1_TRIMER.pdb \n' >> new/tmpk;
printf 'REMARK 250 LOCATION IS CENTERED BETWEEN \n' >> new/tmpk;
printf 'REMARK 250 ALA OXYGEN ATOMS OF RESIDUE 40 IN CHAINS A, E, AND I (TRIMER) \n' >> new/tmpk;
printf 'REMARK 250 AND \n' >> new/tmpk;
printf 'REMARK 250 VAL CG2 ATOMS OF RESIDUE 41 IN CHAINS A, E, AND I (TRIMER) \n' >> new/tmpk;
printf 'HETATM 1 K K A 401 0.000167 35.988 94.21783 K \n' >> new/tmpk;
printf 'END \n' >> new/tmpk;
mv new/tmpk new/figs/fig15/figure_15_panels_G3_G4_4of4-POTASSIUM_ION.pdb;
rm new/tmp.pdb;
rm new/tmpicosframe.pdb;
# (picornavirus_superposition_script) Done.