Weininger Works™ Software
Software Programs
Software Tutorials
Software Output Examples
Release Notes
(WEB PAGE PDF)
Software Programs
chainidPDB, combinePDB, deviation, dist, stripPDB, twwistPDB, and wwavePDB are software programs from Arthur Weininger (www.weiningerworks.com).
- chainidPDB copies an existing PDB file to
a newly created PDB file, replacing one or all chain identifiers with specified chain identifiers.
- combinePDB combines two PDB files into one PDB file.
There are options to specify chain identifier handling.
- deviation reads in multiple files and writes out the population standard deviation
of numerical values that match string token positions across all input files.
Non-numerical values are passed directly to the output as sets of lines (one line
from each file), with numerical values being substituted by the population standard
deviation on the last line of each set of lines. (So if the input files are distance
matrices, then the output file will be a population standard deviation matrix.)
There is an option for producing condensed output (where only the last line of each
set of lines is written to the output), and an option for substituting the mean of
numerical values instead of the population standard deviation.
- dist creates a distance matrix from a points list.
There are options for handling input and output labels and specifying output format.
dist can optionally parse a PDB file, and has options for specifying user defined output atom labels.
- stripPDB removes specified atoms from an input PDB file and writes a new output PDB file.
- twwistPDB transforms the coordinates of one PDB file to the reference frame of another PDB file.
The transformed first PDB file is output as a new PDB file.
The coordinate transformation is specified as 3 specific atoms from the first PDB file
to be respectively mapped to 3 specific atoms of the second reference PDB file.
- wwavePDB identifies atoms in a PDB file within a distance range of other atoms.
wwavePDB is useful for identifying the common spatial occupancy of atoms.
Atom output may be further restricted by set properties (e.g., heterogeneity of chain identifiers, chain identifier content, heterogeneity of atom type)
and individual atom properties (e.g., atom type, atom chain, structural connectivity).
Output is atom-based and is written as PDB output to new PDB files.
wwavePDB identifies:- features without initial target feature input
- contact interface atoms
- binding site features, including between highly divergent structures
- distributed features within a structure or between structures that are not related to contact or binding but represent a structural feature of a group of molecules
- rapidly discover features in a molecular structure or molecular interaction without pre-specifying a search feature
- deliver atom sets for creating common reference surfaces that can be used to compactly display conformational differences between structurally related molecules
- find structural relationships in highly divergent structures
- identify sets of atoms associated with particular molecular functions
- test whether particular common spatial occupancy of atoms in molecules are unique (i.e., not present in other data) and are consistent with their binding without pre-selecting atoms to be considered
Software Tutorials
 |
Picornavirus Monograph Superposition Shell Script: An example of the combined use of chainidPDB, combinePDB, stripPDB, and twwistPDB: (HTML) (PDF) chainidPDB, combinePDB, stripPDB, and twwistPDB are used by the shell script “picornavirus_superposition_script.sh” to create PDB files showing the structural alignments described in the monograph “Common Features in Picornaviruses, Alpha-bungarotoxin, Myelin P2, and CRABP Suggest Structural Bases for Multiple Sclerosis, Guillain-Barré Syndrome, and Paralysis Induction”. |
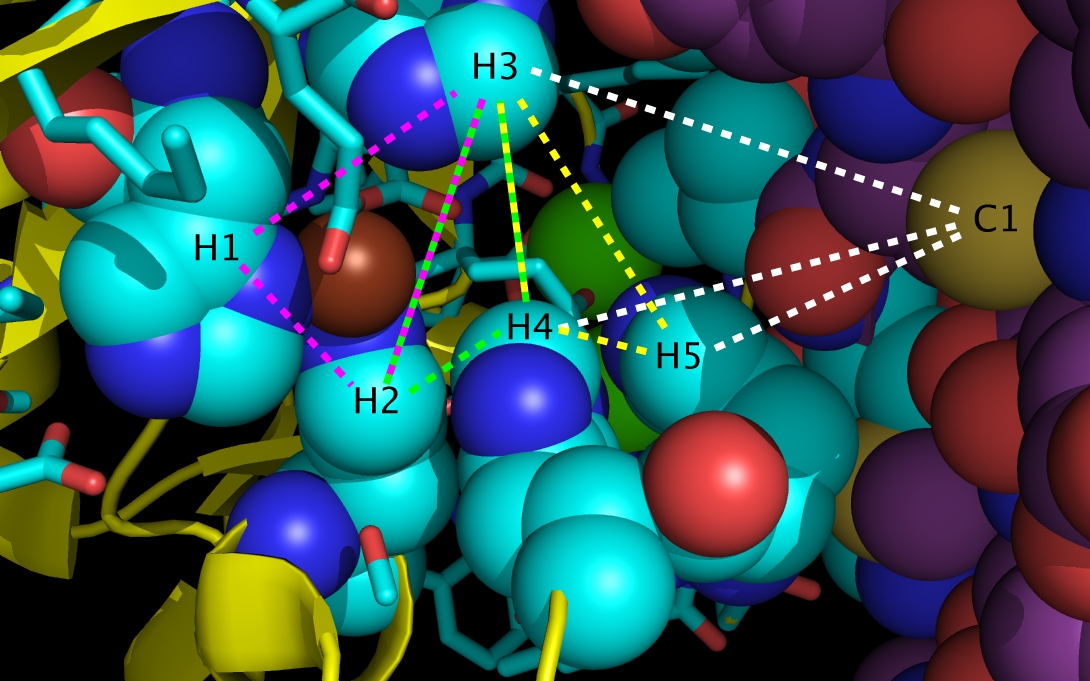 |
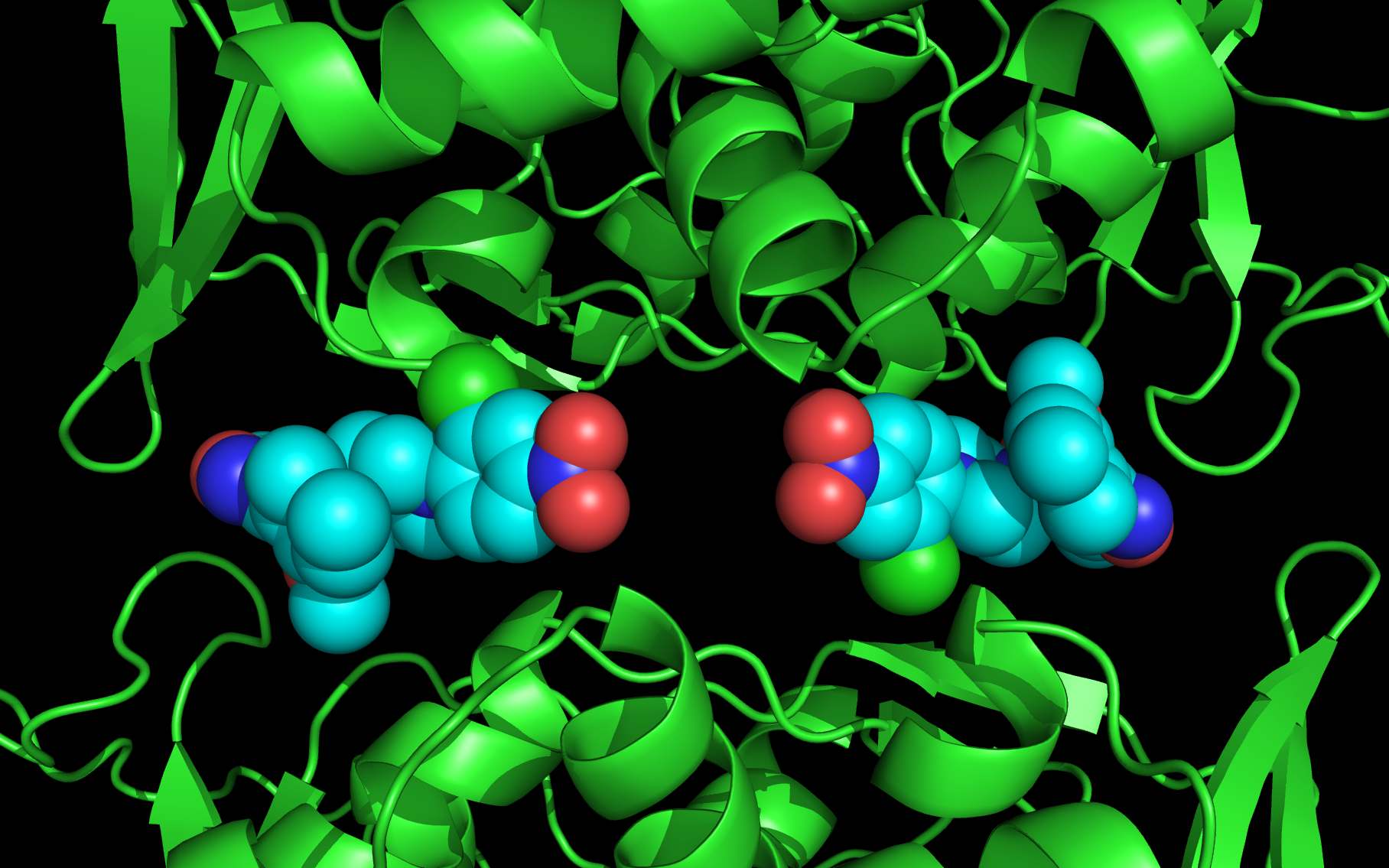
wwavePDB Tutorial #1: Molecular Switch at Sonic Hedgehog/Glycoprotein interface (HTML) (PDF) wwavePDB identifies atoms in a PDB file within a distance range of other atoms. Atom output may be further restricted by set properties (heterogeneity of chain identifiers, chain identifier content, heterogeneity of atom type) and individual atom properties (atom type, atom chain, structural connectivity). This tutorial uses wwavePDB to identify a molecular switch at the interface between sonic hedgehog protein and a bound glycoprotein. This tutorial shows how to use multiple structures in the Protein Data Bank to interpret a specific structure. |
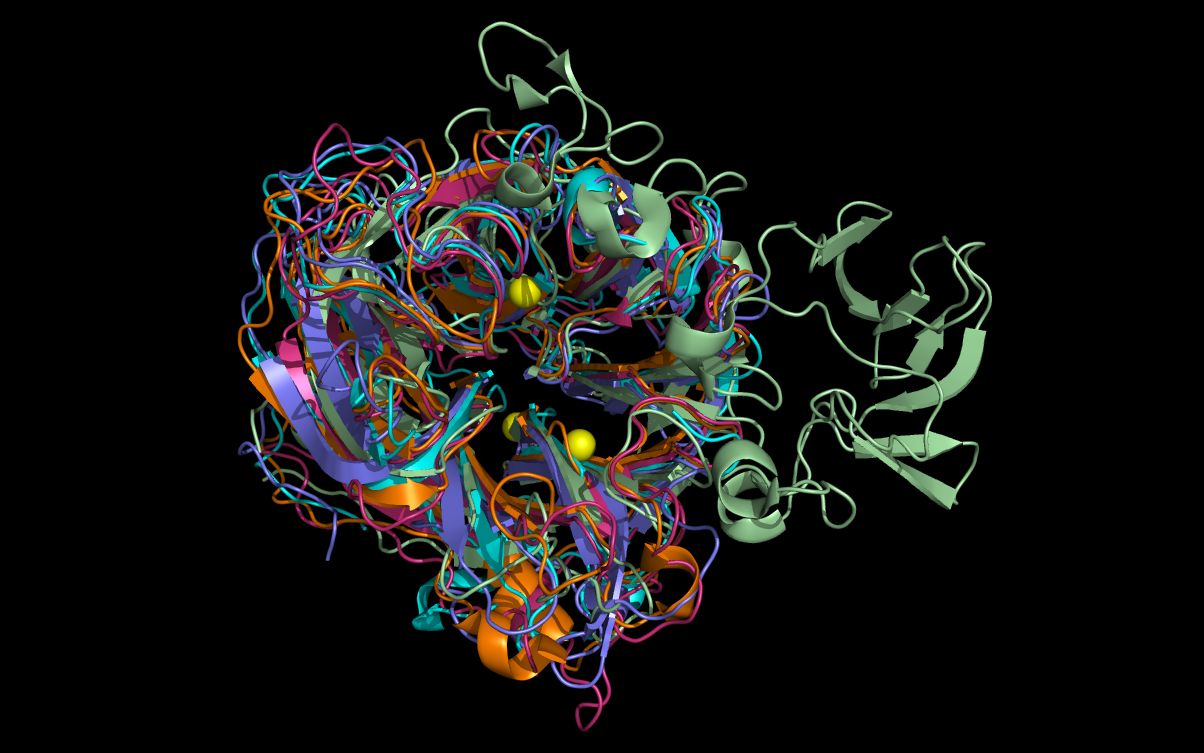 |
twwistPDB Tutorial: Aligning neuraminidase structures and substructures using twwistPDB (HTML) (PDF) twwistPDB transforms the coordinates of one PDB file to the reference frame of another PDB file. This tutorial uses twwistPDB to align neuraminidase structures and substructures. |
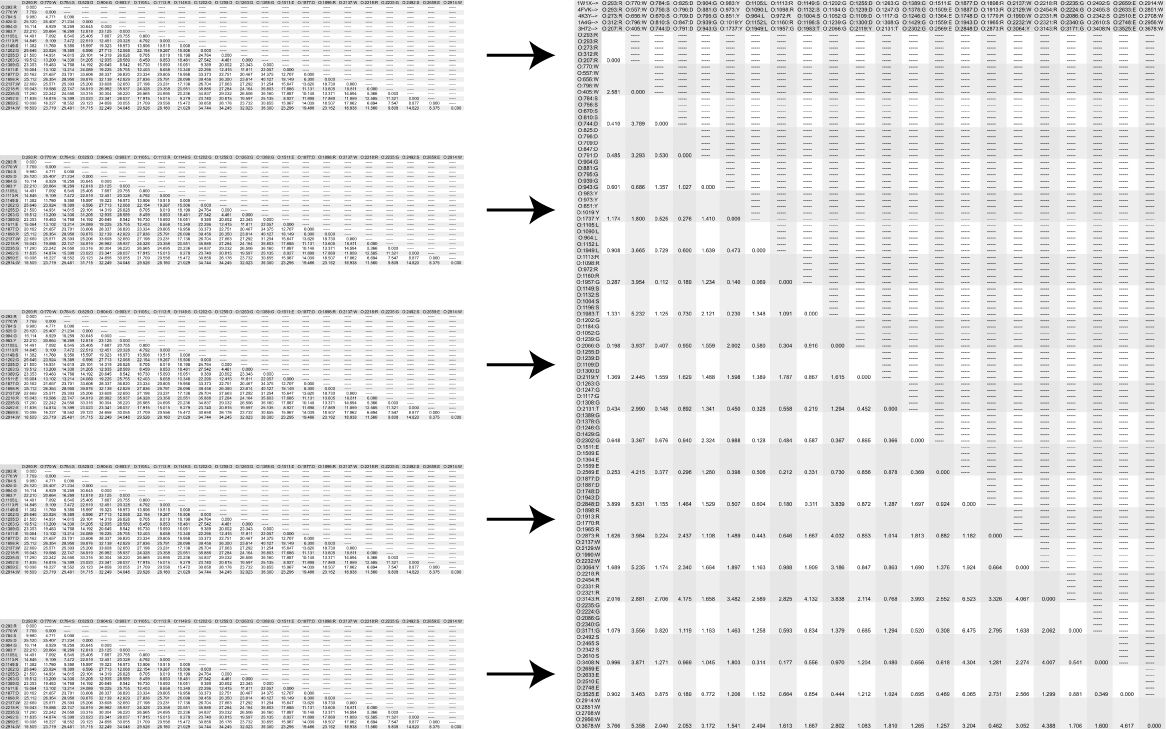 |
dist and deviation Tutorial: Making a population standard deviation matrix for the interatomic distances of 22 specific main chain oxygen atoms in selected neuraminidase proteins. (HTML) (PDF) dist creates a distance matrix from a points list. dist can parse PDB files. deviation reads in multiple files and writes out the population standard deviation of numerical values that match string token positions across all input files. |
Software Output Examples
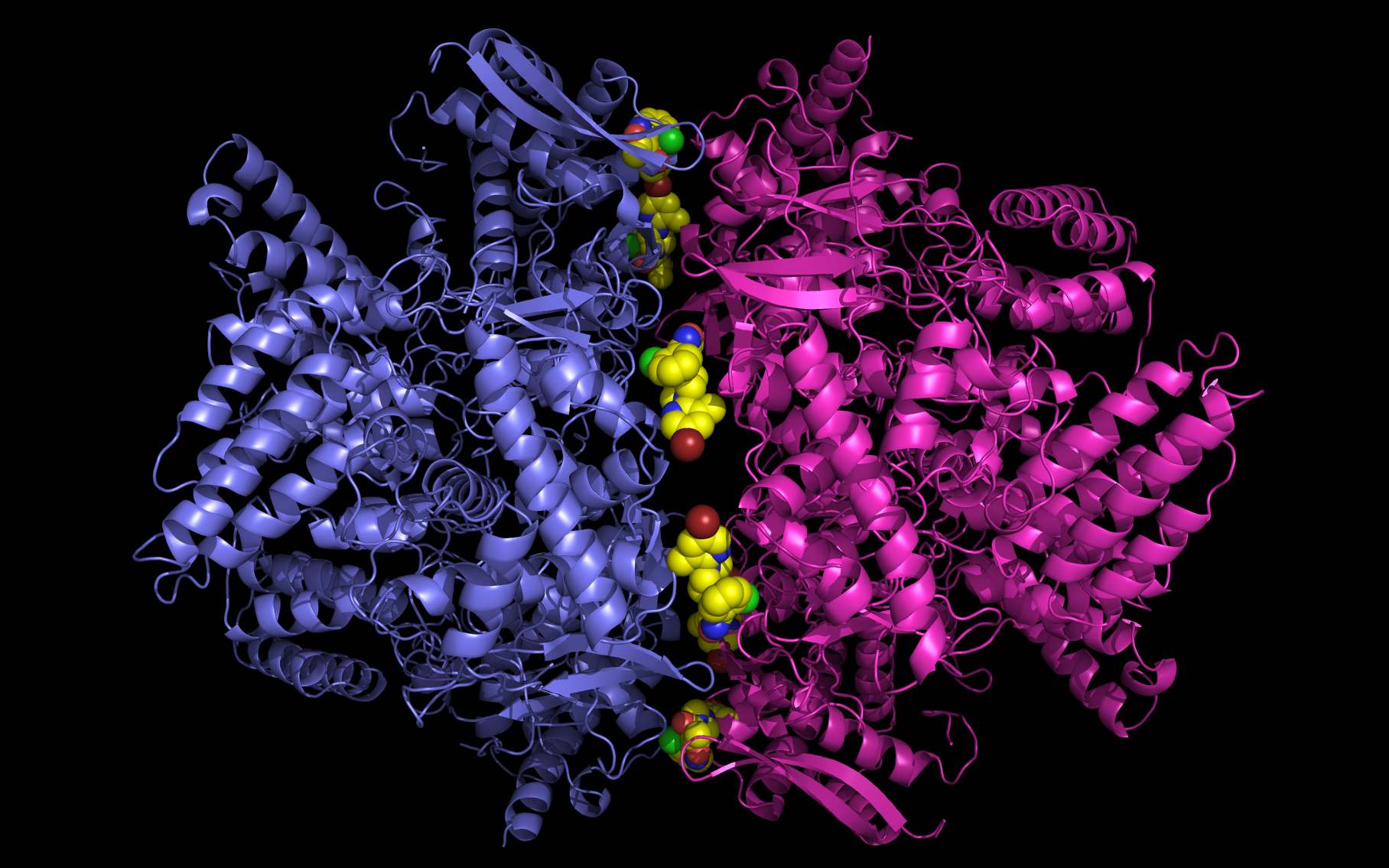 |
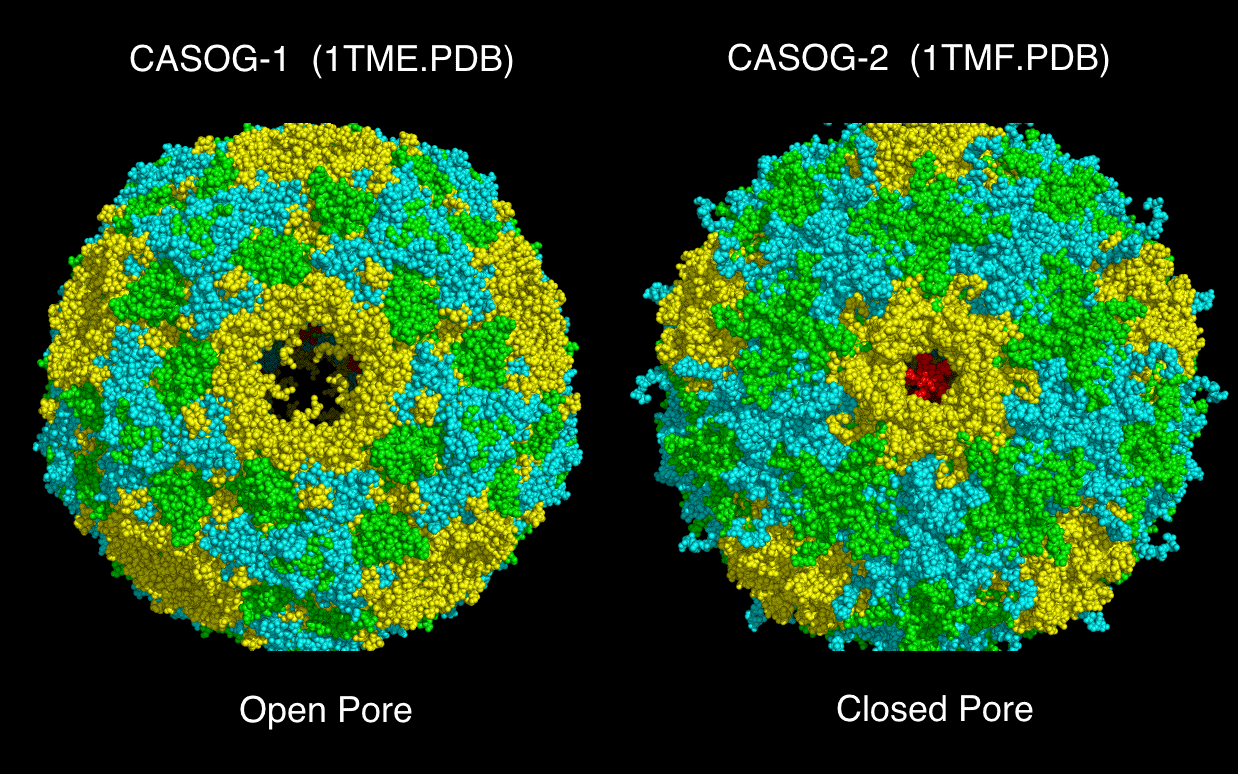
wwavePDB Output Example #4: Finding an alternate binding site in a nucleoprotein trimer-trimer interface (HTML) (PDF) Influenza A nucleoproteins and their bound molecules were analyzed to identify a putative alternative on-target binding site consistent with reported mutational and structural data. |
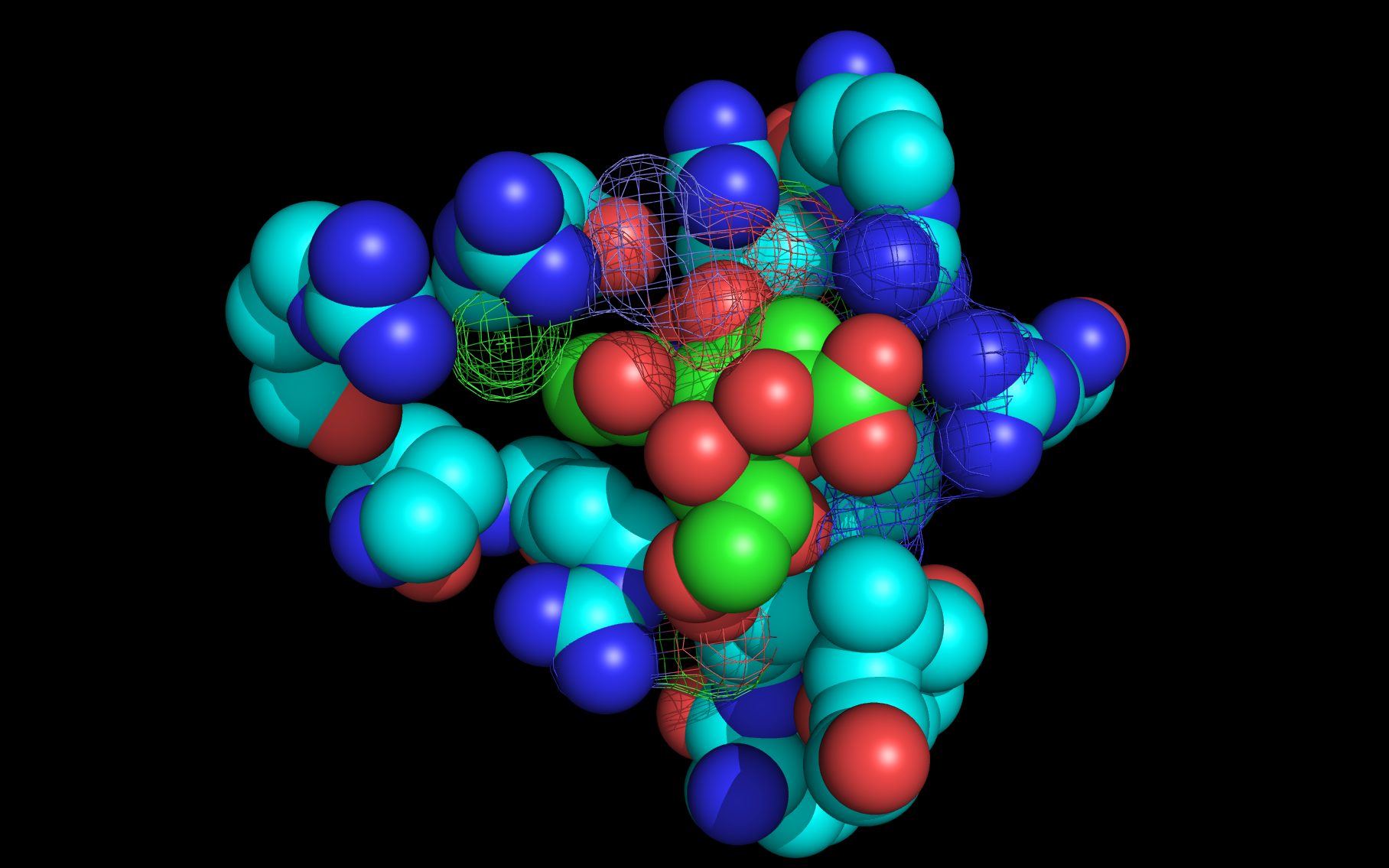 |
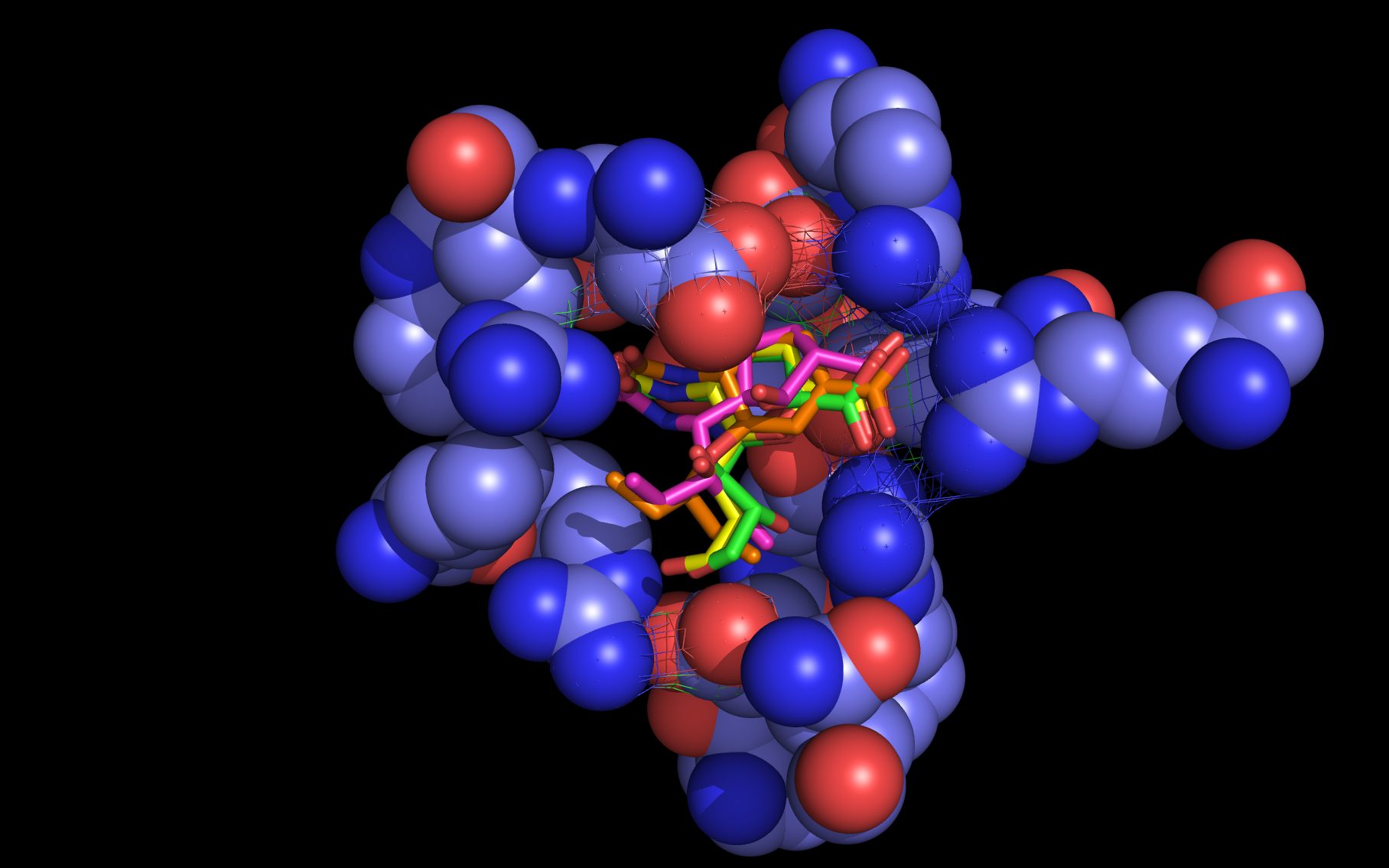
wwavePDB Output Example #3: Finding a sialic acid binding site in a highly divergent N10 Neuraminidase (HTML) (PDF) A putative sialic acid binding site for a highly divergent (~25% sequence identity) bat influenza A virus N10 neuraminidase was identified. A mechanism for enzymatic action was suggested that was based upon common atom presentation in mutated residues and reorientation of residue side chains from their crystal structure positions. |
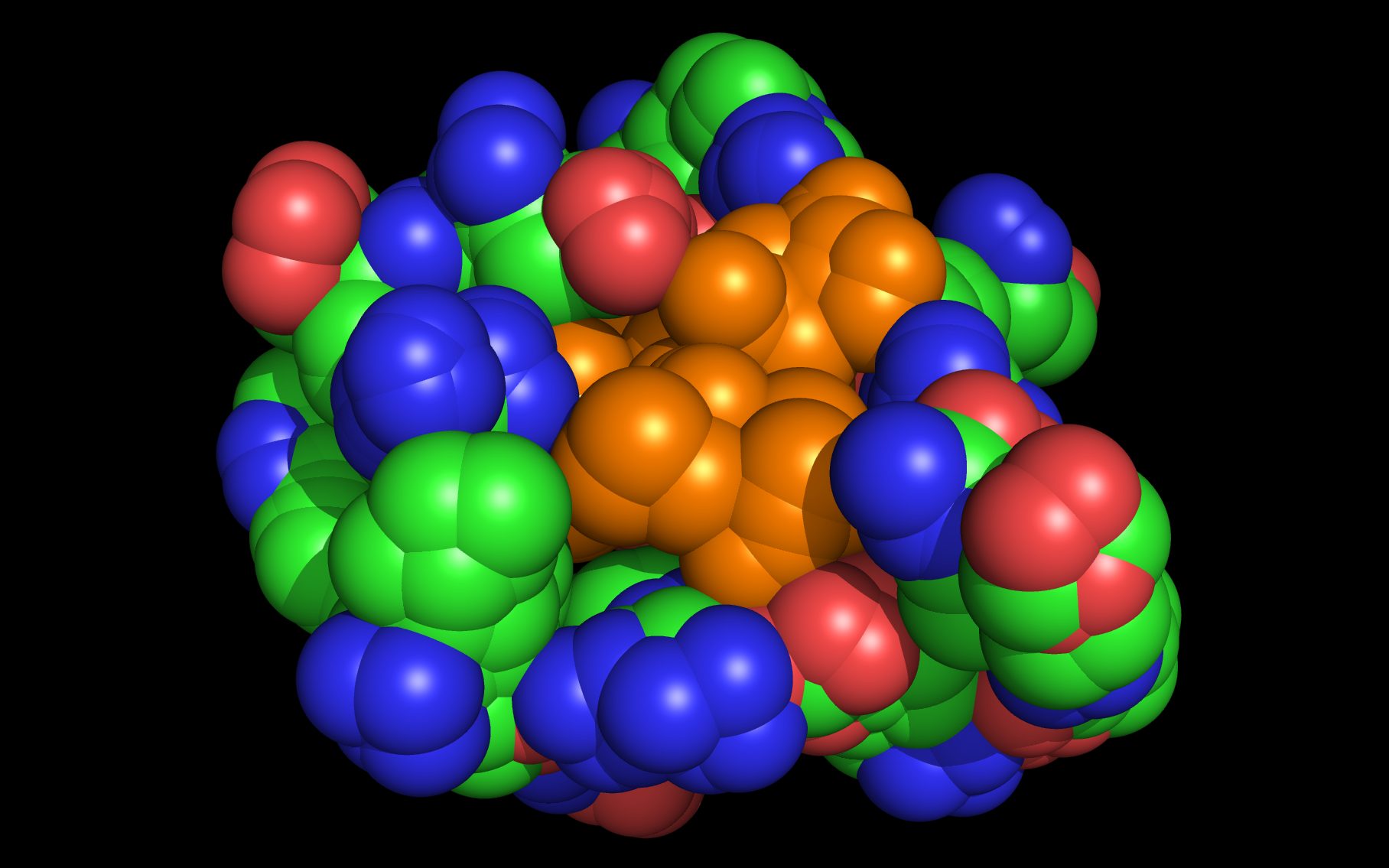 |
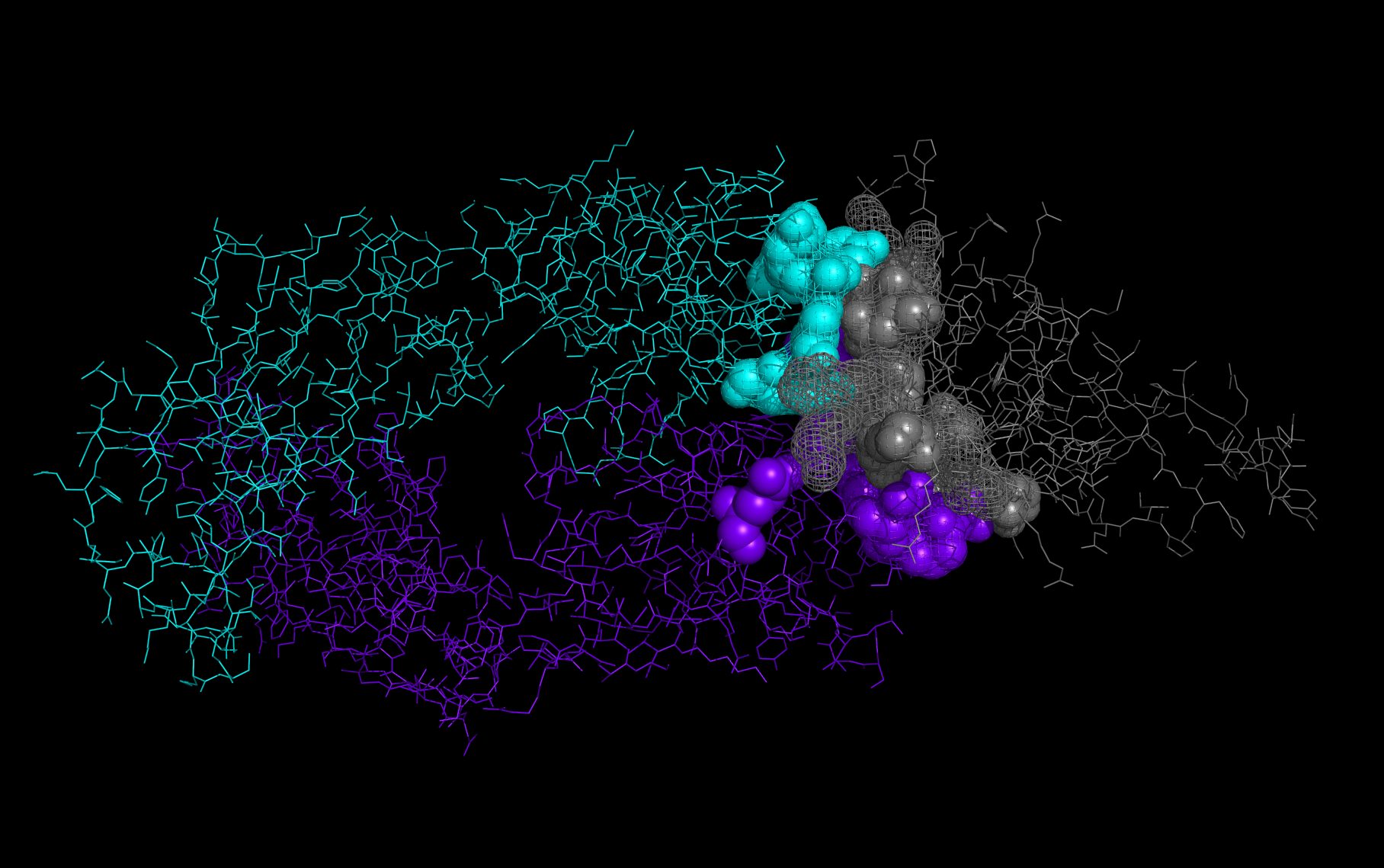
wwavePDB Output Example #2: Using WWaveMarkers and building WWaveCores (HTML) (PDF) wwavePDB generates sets of atoms (“WWaveMarkers”) that can be used to reorient structures into a common reference frame. After structures are reoriented relative to one another, their surfaces can be combined to create common reference surfaces (“WWaveCores”). |
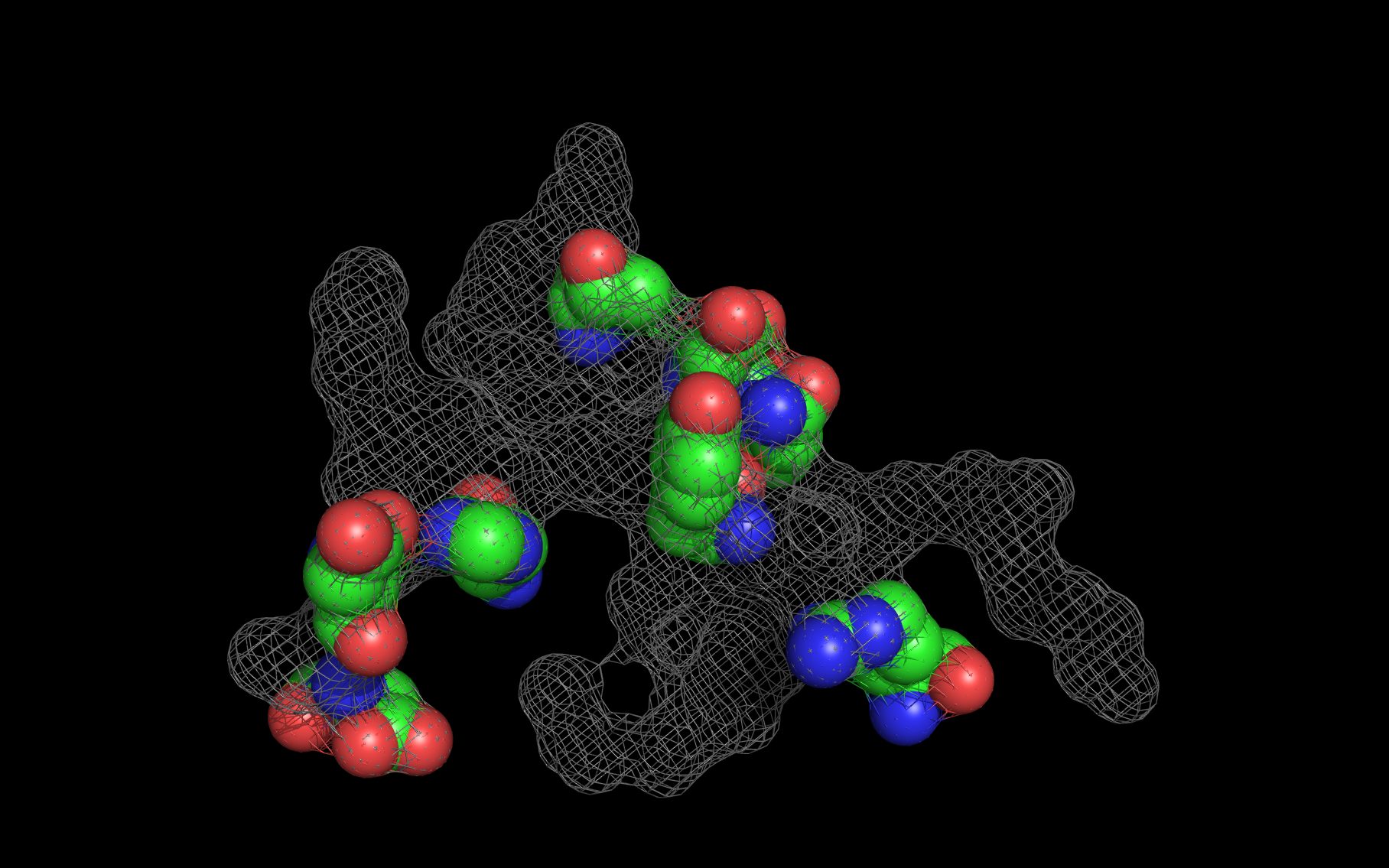 |
wwavePDB Output Example #1: Finding antigen features in an antigen-antibody interface (HTML) (PDF) Basiliximab/IL-2 structure was analyzed in order to show how wwavePDB can be used to identify the contact features of antibody-antigen interfaces. Once the contact features of an antibody-antigen interface are identified, they can be used to determine whether the same features are present elsewhere in molecular databases. |