twwistPDB Tutorial: Aligning neuraminidase structures and substructures
Overview
Example 1: Neuraminidase alignment
Example 2: Staphylococcal Enterotoxin I – N10 neuraminidase substructure alignment
Example 3: Hepatitis E E2s – N11 neuraminidase substructure alignment
Example 4: SARS coronavirus – N11 neuraminidase substructure alignment
Example 5: Alpha-Bungarotoxin – N11 neuraminidase substructure alignment
Example 6: Substance P – N6 neuraminidase substructure alignment
(WEB PAGE PDF)
Tutorial Overview
This tutorial uses twwistPDB to orient sets of pdb files into common alignments. These pdb files have been previously identified by the Weininger Works’ program wwavePDB to contain distributed atoms that have common spatial occupancy (“WWaveMarkers”), i.e. atoms that occupy similar relative positions in space. twwistPDB is used to transform the atom coordinates of a set of pdb files such that these WWaveMarkers are superimposed.
twwistPDB Overview Page describes twwistPDB command line syntax.
Example 1 aligns the neuraminidases from the set of the following pdb files:
- 1W1X: N6 (influenza A) (PDB) (fasta) (summary)
- 4FVK: N10 (influenza A H17N10) (PDB) (fasta) (summary)
- 4K3Y: N11 (influenza A H18N11) (PDB) (fasta) (summary)
- 1A4G: INB (influenza B) (PDB) (fasta) (summary)
- 3H72: Streptococcus pneumoniae R6 (PDB) (fasta) (summary)
Example 2 aligns specific staphylococcal enterotoxin I (“SEI”) residues (from 2G9H) and N10 neuraminidase residues (from 4FVK):
- 2G9H: staphylococcal enterotoxin I (SEI) (PDB) (fasta) (summary)
- 4FVK: N10 (influenza A H17N10) (PDB) (fasta) (summary)
Example 3 aligns specific hepatitis E E2s residues (from 3RKD) and N11 neuraminidase residues (from 4K3Y):
- 3RKD: hepatitis E virus E2s (PDB) (fasta) (summary)
- 4K3Y: N11 (influenza A H18N11) (PDB) (fasta) (summary)
Example 4 aligns specific SARS coronavirus spike protein receptor-binding-like residues (from 3SCK) and N11 neuraminidase residues (from 4K3Y):
- 3SCK: SARS coronavirus (PDB) (fasta) (summary)
- 4K3Y: N11 (influenza A H18N11) (PDB) (fasta) (summary)
Example 5 aligns specific alpha-bungarotoxin residues (from 2ABX) and N11 neuraminidase residues (from 4K3Y):
- 2ABX: alpha-bungarotoxin (PDB) (fasta) (summary)
- 4K3Y: N11 (influenza A H18N11) (PDB) (fasta) (summary)
Example 6 aligns specific substance P residues (from 2KS9) and N6 neuraminidase residues (from 1W1X):
- 2KS9: Substance P (PDB) (fasta) (summary)
- 1W1X: N6 (influenza A) (PDB) (fasta) (summary)
- WSUBP.pdb (substance P with side chains reoriented to match the N6 neuraminidase) (PDB)
Example 1: Neuraminidase alignment
Example 1: Overview
Specific neuraminidases (N6 (1W1X), N10 (4FVK), N11 (4K3Y), influenza B = “INB” (1A4G), and streptococcus = “STR” (3H72)) contain distributed atoms that have common spatial occupancy (“WWaveMarkers”), i.e. atoms that occupy similar relative positions in space. We refer to these WWaveMarkers with a common numbering system (“CNS”): CNS 118:O, CNS 224:O and CNS 276:O. In the published structures, these atoms do not occupy the same coordinates. twwistPDB is used to transform the atom coordinates of a set of pdb files such that these WWaveMarkers, and their relative N6, N10, N11, INB, and STR structures, are superimposed.
The WWaveMarkers for the N6, N10, N11, INB and STR neuraminidase structures are:
| PDB | N.Type | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 1W1X | N6 | B | 1124 | ARG | 3302 | O |
| 4FVK | N10 | B | 118 | ARG | 3169 | O |
| 4K3Y | N11 | A | 118 | ARG | 273 | O |
| 1A4G | INB | A | 115 | ARG | 312 | O |
| 3H72 | STR | A | 347 | ARG | 207 | O |
| PDB | N.Type | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 1W1X | N6 | B | 1231 | ARG | 4122 | O |
| 4FVK | N10 | B | 224 | ARG | 3974 | O |
| 4K3Y | N11 | A | 224 | ARG | 972 | O |
| 1A4G | INB | A | 222 | ARG | 1160 | O |
| 3H72 | STR | A | 567 | GLY | 1957 | O |
| PDB | N.Type | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 1W1X | N6 | B | 1283 | GLU | 4520 | O |
| 4FVK | N10 | B | 276 | GLU | 4385 | O |
| 4K3Y | N11 | A | 276 | GLU | 1364 | O |
| 1A4G | INB | A | 274 | GLU | 1559 | O |
| 3H72 | STR | A | 647 | GLU | 2569 | O |
Example 1: Instructions
- Request and install twwistPDB
(if you have not already done so).
https://weiningerworks.com/contact.html - Download PDB structure files from rcsb.org (if you have not already done so):
1W1X, 4FVK, 4K3Y, 1A4G, 3H72
- Run twwistPDB to orient pdb files to one arbitrary pdb file (1W1X.pdb in the following).
twwistPDB will output a single reoriented pdb file (N10ontoN6.pdb, N11ontoN6.pdb, INBontoN6.pdb, and STRontoN6.pdb in the following) for each twwistPDB execution.
(Note: as of version 1.0.1, twwistPDB automatically alters the rotation/translation order to minimize alignment error.)
(Note: “.ent” files can be used instead of “.pdb” files.)
cp 1W1X.pdb N6.pdb
twwistPDB -o N10ontoN6.pdb 4FVK.pdb 3169 3974 4385 1W1X.pdb 3302 4122 4520
twwistPDB -o N11ontoN6.pdb 4K3Y.pdb 273 972 1364 1W1X.pdb 3302 4122 4520
twwistPDB -o INBontoN6.pdb 1A4G.pdb 312 1160 1559 1W1X.pdb 3302 4122 4520
twwistPDB -o STRontoN6.pdb 3H72.pdb 207 1957 2569 1W1X.pdb 3302 4122 4520
- The four new pdb files containing “ontoN6” all share the same orientation as 1W1X.pdb (“N6.pdb”).
Open the 5 files containing “N6” at the same time in a molecular visualization program. Note that the twwistPDB commands superimposed N10, N11, INB, and STR structures with the N6 structure despite low (N10, N11 and INB) or absent (STR) sequence homology with the N6 structure.
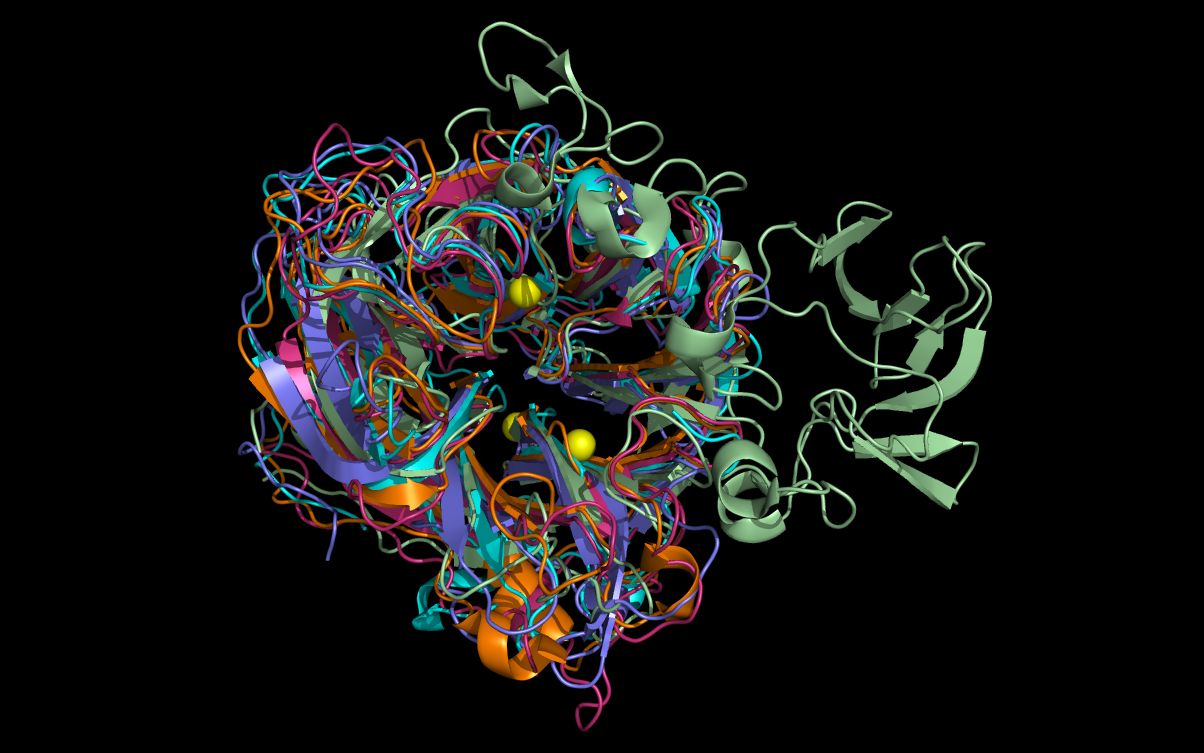
The alignment of putative active site residues in the N10 and N11 structures have not been optimized as the main and side chain atoms have not been moved. These side chain residues can be moved from the superimposed crystal structure positions to achieve a better residue superposition as illustrated for the N10 neuraminidase in this HTML page or in this downloadable PDF file. (In the HTML, mouse over the small index images below the main top image to change that larger image and associated description.)
Example 2: Staphylococcal Enterotoxin I – N10 neuraminidase substructure alignment
Example 2: Overview
Certain staphylococcal enterotoxin I (“SEI”) (2G9H) and N10 neuraminidase (4FVK) residues have common spatial occupancy, i.e. their atoms occupy a similar volume. The program wwavePDB identified specific distributed atoms for SEI and N10 having common spatial occupancy (“WWaveMarkers”), i.e. atoms that occupy similar relative positions in space. In the published structures, these atoms do not occupy the same coordinates. twwistPDB is used to transform SEI atom coordinates to be superimposed over the N10 atom coordinates. No relative position within a structure is changed.
The WWaveMarkers for the SEI and N10 are:
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4FVK | A | 106 | TRP | 207 | NE1 |
| 2G9H | D | 51 | TRP | 3509 | NE1 |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4FVK | A | 109 | GLU | 225 | CA |
| 2G9H | D | 53 | GLU | 3524 | CA |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4FVK | A | 139 | SER | 465 | O |
| 2G9H | D | 34 | GLN | 3373 | O |
Example 2: Instructions
- Request and install twwistPDB
(if you have not already done so).
https://weiningerworks.com/contact.html - Download PDB structure files from rcsb.org (if you have not already done so):
2G9H, 4FVK
- Run twwistPDB to orient 2G9H.pdb to 4FVK.pdb.
twwistPDB will output a single reoriented pdb file, SEIontoN10.pdb.
cp 4FVK.pdb N10.pdb
twwistPDB -o SEIontoN10.pdb 2G9H.pdb 3509 3524 3373 4FVK.pdb 207 225 465
- The new pdb file, SEIontoN10.pdb, shares the same orientation as 4FVK.pdb (“N10.pdb”).
Open SEIontoN10.pdb and N10.pdb at the same time in a molecular visualization program. Note that the twwistPDB command superimposed common staphylococcal enterotoxin I and N10 neuraminidase residues.
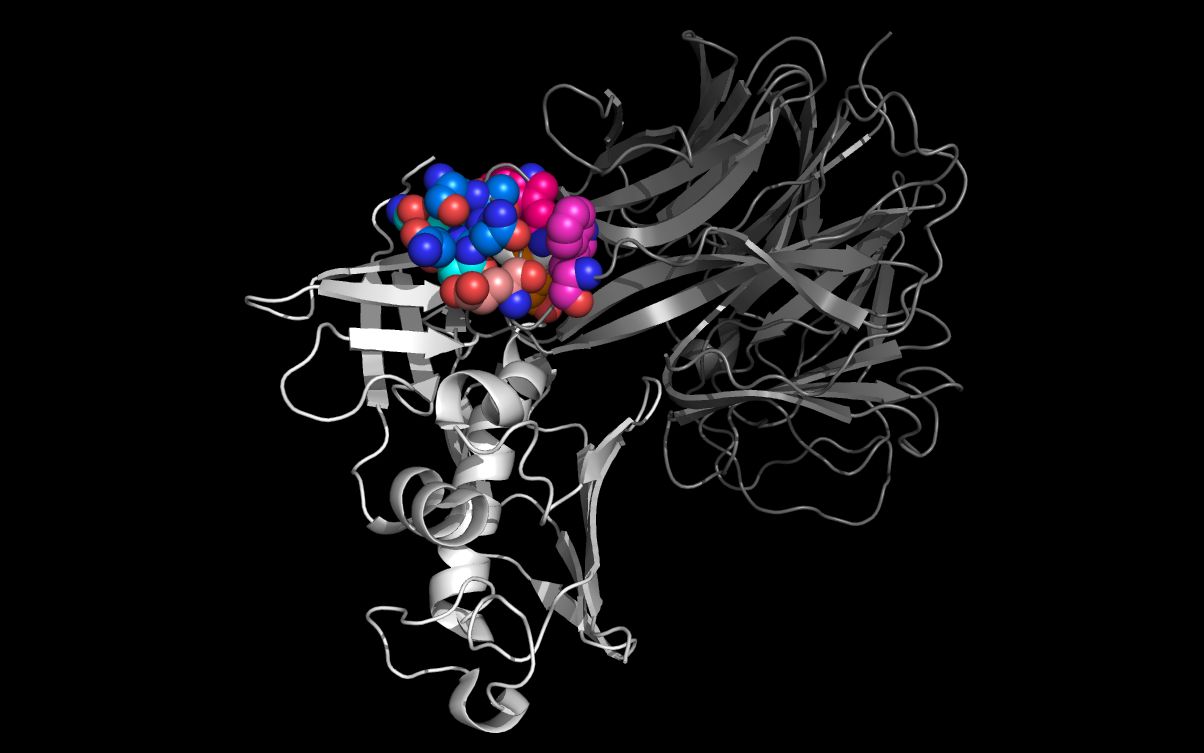
The picture above shows:- SEI monomer ribbon colored white
- SEI residue S31, A32, N33, Q34 carbon spheres colored green
- SEI residue W51 and E53 carbon spheres colored red
- N10 neuraminidase monomer cartoon colored blue
- N10 neuraminidase tetramer WWaveMarkers (lines and residue #) colored blue
- N10 neuraminidase monomer residue S139, A140, N141, Q142 carbon spheres colored orange
- N10 neuraminidase monomer residue W106 and E109 carbon spheres colored yellow
The alignment of putative cell entry domain residues in the staphylococcal enterotoxin I (2G9H residues S31, A32, N33, Q34, W51, and E53) and on the N10 residues on the surface of the H17N10 virion (4FVK residues S139, A140, N141, Q142, W106, and E109) have not been optimized as the main and side chain atoms have not been moved. These side chain residues can be moved from the superimposed crystal structure positions to achieve a better residue superposition.
Example 3: Hepatitis E E2s – N11 neuraminidase substructure alignment
Example 3: Overview
Certain hepatitis E E2s (3RKD) and N11 neuraminidase (4K3Y) residues have common spatial occupancy, i.e. their atoms occupy a similar volume. The program wwavePDB identified specific distributed atoms for hepatitis E E2s and N11 having common spatial occupancy (“WWaveMarkers”), i.e. atoms that occupy similar relative positions in space. In the published structures, these atoms do not occupy the same coordinates. twwistPDB is used to superimpose hepatitis E E2s atom coordinates over the N11 atom coordinates. No relative position within a structure is changed.
The WWaveMarkers for the hepatitis E E2s and N11 are:
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 138 | TYR | 439 | CB |
| 3RKD | A | 557 | TYR | 754 | CB |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | C | 431 | LYS | 8001 | CA |
| 3RKD | A | 554 | LYS | 733 | CA |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | C | 430 | THR | 7997 | CB |
| 3RKD | A | 553 | THR | 729 | CB |
Example 3: Instructions
- Request and install twwistPDB
(if you have not already done so).
https://weiningerworks.com/contact.html - Download PDB structure files from rcsb.org (if you have not already done so):
3RKD, 4K3Y
- Run twwistPDB to orient 3RKD.pdb to 4K3Y.pdb.
twwistPDB will output a single reoriented pdb file, H2SontoN11.pdb.
cp 4K3Y.pdb N11.pdb
twwistPDB -o H2SontoN11.pdb 3RKD.pdb 754 733 729 4K3Y.pdb 439 8001 7997
- The new pdb file, H2SontoN11.pdb, shares the same orientation as 4K3Y.pdb (“N11.pdb”).
Open H2SontoN11.pdb and N11.pdb at the same time in a molecular visualization program. Note that the twwistPDB command superimposed common hepatitis E2S and N11 neuraminidase residues.
The alignment of putative cell entry domain residues in the hepatitis E2S residues (3RKD residues G551, T552, T553, K554, A555, G 556, Y557, Y561) and on the N11 residues on the surface of the H18N11 virion (4K3Y residues A428, T429, T430, K431, A432, G433, Y138, Y159) have not been optimized as the main and side chain atoms have not been moved. These side chain residues can be moved from the superimposed crystal structure positions to achieve a better residue superposition.
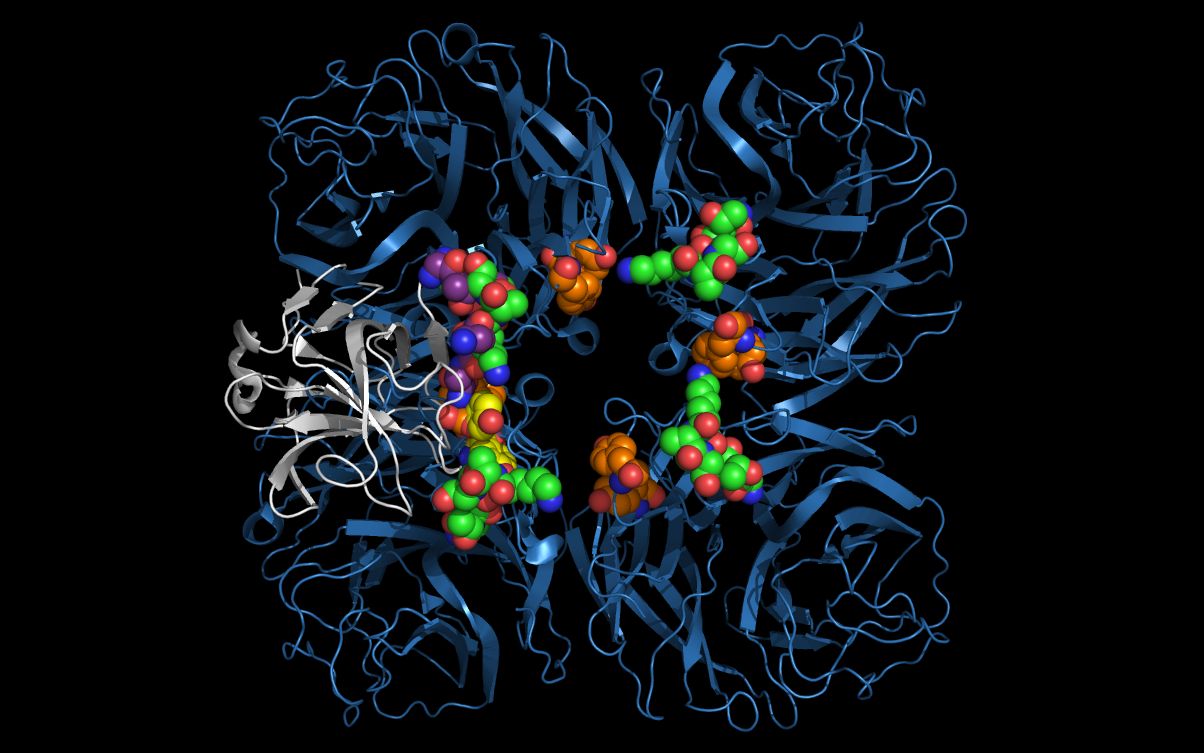
The picture above shows:- N11 first neuraminidase tetramer cartoon colored blue
- Hepatitis E E2S protein monomer cartoon colored white
- N11 neuraminidase tetramer residue T430, K431, A432, G433 carbon spheres colored green
- Hepatitis E E2S residue T553, K554, A555, G556 carbon spheres colored purple
- N11 neuraminidase tetramer residue Y138 and Y159 carbon spheres are colored orange
- Hepatitis E E2S residue Y557 and Y561 carbon spheres colored yellow
The hepatitis E2S residues (3RKD residues G551, T552, T553, K554, A555, G556, Y557, Y561) are optimally mapped onto residues in the N11 tetramer from two N11 monomers: G551, T552, T553, K554, A555, and G556 in one monomer and Y138 and Y159 in another monomer, i.e., residues comprising the putative hepatitis E2S cell entry domain are distributed across monomers in the N11 tetramer.
Example 4: SARS coronavirus – N11 neuraminidase substructure alignment
Example 4: Overview
Certain SARS spike protein (“SARSP”) residues (3SCK) and N11 neuraminidase residues (4K3Y) have common spatial occupancy, i.e. their atoms occupy a similar volume. The program wwavePDB identified specific distributed atoms for SARS spike protein residues and N11 having common spatial occupancy (“WWaveMarkers”), i.e. atoms that occupy similar relative positions in space. In the published structures, these atoms do not occupy the same coordinates and there are several sets of atoms on mobile loops. Domain 1 in SARSP (“SARSPD1”) contains residues T485, T486, T487, and G488. The corresponding domain 1 in N11 neuraminidase (“N11D1”) contains residues G105, T106, T107, and G108. Domain 2 in SARSP (“SARSPD2”) contains residues P469, P470, A471, P472. The corresponding domain 2 in N11 neuraminidase (“N11D2”) contains residues P166, P167, T168 and P169. Domain 3 in SARSP (“SARSPD3”) contains residues T425, R426, and N427. The corresponding domain 3 residues in N11 neuraminidase (“N11D3”) are N401, S402, and T403.
twwistPDB is used to superimpose SARS coronavirus atom coordinates over the N11 atom coordinates. No relative position within a structure is changed. A set of three SARSP atoms (3SCK) are superimposed on the set of N11 neuraminidase atoms (4K3Y) so that similar presentation of the atoms by the structures can be assessed.
The WWaveMarkers for the SARSP residues and N11 are:
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | C | 107 | THR | 5637 | O |
| 3SCK | F | 486 | THR | 12385 | O |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | C | 403 | THR | 7764 | O |
| 3SCK | F | 425 | THR | 11880 | O |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | D | 167 | PRO | 8738 | N |
| 3SCK | F | 470 | PRO | 12247 | N |
Example 4: Instructions
- Request and install twwistPDB
(if you have not already done so).
https://weiningerworks.com/contact.html - Download PDB structure files from rcsb.org (if you have not already done so):
3SCK, 4K3Y
- Run twwistPDB to orient 3SCK.pdb to 4K3Y.pdb.
twwistPDB will output a single reoriented pdb file, SARSPontoN11.pdb.
cp 4K3Y.pdb N11.pdb
twwistPDB -o SARSPontoN11.pdb 3SCK.pdb 12385 11880 12247 4K3Y.pdb 5637 7764 8738
- The new pdb file, SARSPontoN11.pdb, shares the same orientation as 4K3Y.pdb (“N11.pdb”).
Open SARSPontoN11.pdb and N11.pdb at the same time in a molecular visualization program. Note that the twwistPDB command superimposed common SARSPontoN11 residues and N11 neuraminidase residues.
In order to relate SARSP domains to N11 neuraminidase domains, twwistPDB was used to reorient SARSP residue T486, T425 and P470 atoms in chain “F” onto N11 neuraminidase residue atoms in T107 and T403 in chain “C” and in P167 in chain “D”. This results in domains in the SARSP molecule being mapped onto domains in the N11 neuraminidase tetramer with the same relative orientation without changing the relative crystal structure positions of either the SARSP or the N11 neuraminidase atoms. The loop containing residues P105-P108 in the N11 neuraminidase and residues P469-P472 in the SARSP is mobile and, if moved, could also reach the corresponding residues within the monomer, instead of (as crystallized) across monomers in the tetramer.
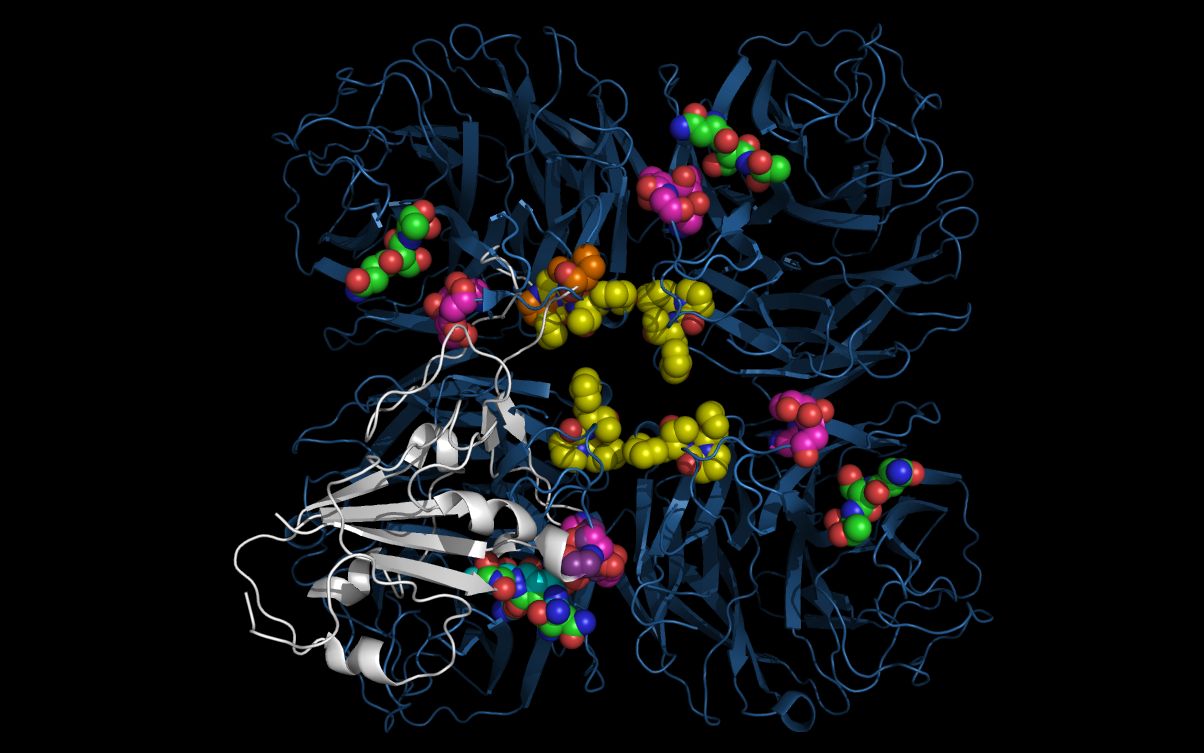
The picture above shows:
- N11 tetramer ribbons, colored blue
- SARSP monomer ribbon, colored white, in the lower left quadrant
- N11 domain 1 residue carbon spheres colored magenta
- N11 domain 2 residue carbon spheres colored yellow
- SARSP domain 1 residue carbon spheres colored purple
- SARSP domain 2 residue carbon spheres colored orange
- N11 domain 3 and SARSP domain 3 residue carbon spheres colored green
Although the SARSP is reoriented by twwistPDB onto the N11 monomer so that SARSP domain 2 is in a different monomer (upper left quadrant) than SARSP domain 1 (lower left quadrant), the loop containing SARSP domain 2 is flexible and the SARSP domain 2 residues (carbon spheres colored orange) can be repositioned over the N11 lower left quadrant N11 domain 2 residues (carbon spheres colored yellow). This loop repositioning would overlap SARSP monomer domains 1, 2, and 3 within a single N11 monomer.
Example 5: Alpha-Bungarotoxin – N11 neuraminidase substructure alignment
Example 5: Overview
Certain alpha-bungarotoxin (“ABT”) residues (2ABX) and N11 neuraminidase (4K3Y) residues have common spatial occupancy, i.e. their atoms occupy a similar volume. The program wwavePDB identified specific distributed atoms for alpha-bungarotoxin residues and N11 having common spatial occupancy (“WWaveMarkers”), i.e. atoms that occupy similar relative positions in space. In the published structures, these atoms do not occupy the same coordinates. There are two sets of atoms (domains 1 and 2) on mobile loops. Domain 1 in ABT (“ABTD1”) contains residues T62, D36 and K64. Domain 1 in N11 neuraminidase (“N11D1”) contains residues S126, D126 and K127. Domain 2 in ABT (“ABTD2”) contains residues G19, E20, N21, L22 and C23. Domain 2 in N11 neuraminidase (“N11D2”) contains residues G88, E89, N90, L91 and C92.
In order to reorient two alpha-bungarotoxin (“ABT”) domains to two N11 neuraminidase domains, twwistPDB was used to superimpose N11 neuraminidase residues D126 and K127 onto ABT residues D63 and K64, and to superimpose N11 neuraminidase residues E89 and N90 onto ABT residues E20 and N21. This results in domain 1 residues and domain 2 residues from the two sets of N11 neuraminidase monomers being mapped onto alpha-bungarotoxin in the same relative orientation without changing the relative crystal structure positions of N11 neuraminidase atoms. This could also be accomplished by changing the relative (main chain loop and side chain) positions of the N-terminal residues of N11 so that N11D2 neuraminidase matched the ABTD2 alpha-bungarotoxin atom positions in a single monomer. No relative position within a structure is changed.
The first set of three N11D1 atoms (4K3Y) are superimposed on the set of ABT (2ABX) atoms so that similar presentation of the atoms by the structures can be assessed.
The WWaveMarkers for the first set of alpha-bungarotoxin residues and N11 are:
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 126 | ASP | 337 | O |
| 2ABX | A | 63 | ASP | 459 | O |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 126 | ASP | 340 | OD2 |
| 2ABX | A | 63 | ASP | 462 | OD1 |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 127 | LYS | 348 | CE |
| 2ABX | A | 64 | LYS | 470 | CE |
The WWaveMarkers for the second set of alpha-bungarotoxin residues and N11 are:
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 89 | GLU | 52 | CG |
| 2ABX | A | 20 | GLU | 133 | CG |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 90 | ASN | 57 | CA |
| 2ABX | A | 21 | ASN | 138 | CA |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 4K3Y | A | 90 | ASN | 60 | CB |
| 2ABX | A | 21 | ASN | 141 | CB |
Example 5: Instructions
- Request and install twwistPDB
(if you have not already done so).
https://weiningerworks.com/contact.html - Download PDB structure files from rcsb.org (if you have not already done so):
2ABX, 4K3Y
- Run twwistPDB to orient 4K3Y.pdb to 2ABX.pdb.
twwistPDB will output a single reoriented pdb file (N11D1onABT.pdb, N11D2onABT.pdb in the following) for each twwistPDB execution.
cp 2ABX.pdb ABT.pdb
twwistPDB -o N11D1onABT.pdb 4K3Y.pdb 337 340 348 2ABX.pdb 459 462 470
twwistPDB -o N11D2onABT.pdb 4K3Y.pdb 52 57 60 2ABX.pdb 133 138 141
- The alignment of N11 neuraminidase domains 1 and 2 onto acetylcholine binding residues
in alpha-bungarotoxin produces substantially the same combined domain as in alpha-bungarotoxin.
Open N11D1onABT.pdb, N11D2onABT.pdb, and ABT.pdb at the same time in a molecular visualization program.
For the best comparison of the translocated domains, display residues S93-G459 of N11D1onABT.pdb and residues 82-92 of N11D1onABT.pdb. The structure formed from N11 residues S126, D126, K127 (N11D1) and residues G88, E89, N90, L91 and C92 (N11D2) can be compared to ABT residues T62, D36 and K64 (ABTD1) residues G19, E20, N21, L22 and C23 (ABTD2).
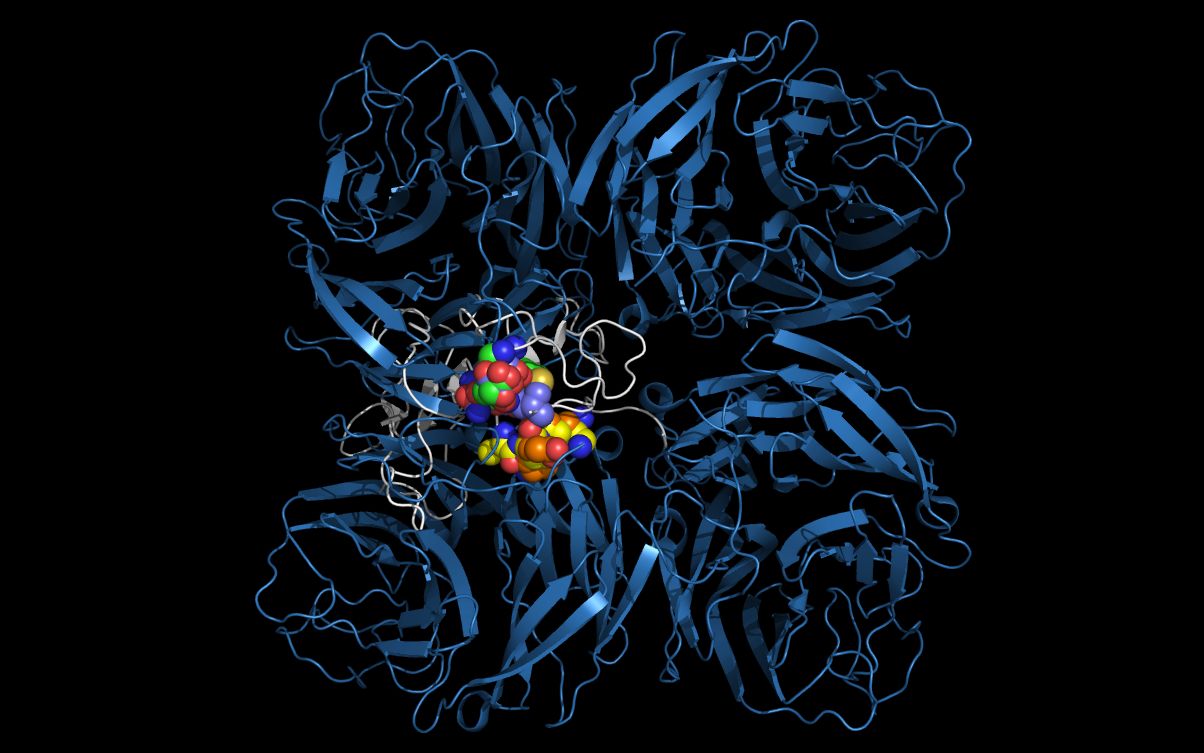
The picture above shows:- N11 first neuraminidase tetramer cartoon colored blue
- Alpha-bungarotoxin monomer cartoon colored white
- N11 first tetramer residue G88, E89, N90, L91, C92 carbon spheres colored purple
- Alpha-bungarotoxin residue G19, E20, N21, L22, C23 carbon spheres colored green
- N11 second tetramer residue S125, D126, K127 carbon spheres colored orange
- Alpha-bungarotoxin residue T62, D63, K64 carbon spheres colored yellow
Using twwistPDB to reposition two N11 tetramers repositions the residues onto the alpha-bungarotoxin without loop repositioning. Repositioning of the two tetramers places the groups in the same position as if the loops were moved and allows the evaluation, without loop rebuilding, of the ability of an N11 neuraminidase monomer to form an alpha-bungarotoxin cell entry domain.
Example 6: Substance P – N6 neuraminidase substructure alignment
Example 6: Overview
Certain substance P (2KS9) and N6 neuraminidase (1W1X) residues have common spatial occupancy, i.e. their atoms occupy a similar volume. In the published structures, these atoms do not occupy the same coordinates. The small (11 amino acids) substance P is highly flexible and multiple N-terminal conformers occupy the same volume. In order to map substance P domains to N6 neuraminidase domains, substance P residues R365, P366, and K367 (the first, second, and third residues of substance P) have been reoriented to place the substance P atoms in the same configuration as the same residues in the N6 neuraminidase. twwistPDB is used to superimpose substance P atom coordinates over the N6 atom coordinates.
The WWaveMarkers for substance P and N6 are:
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 1W1X | C | 2438 | ARG | 8715 | CA |
| WSUBP | B | 365 | ARG | 5869 | CA |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 1W1X | C | 2439 | PRO | 8726 | CA |
| WSUBP | B | 366 | PRO | 5895 | CA |
| PDB | Chain | Residue # | Residue Type | Atom # | Atom Name |
| 1W1X | C | 2440 | LYS | 8733 | CA |
| WSUBP | B | 367 | LYS | 5909 | CA |
Example 6: Instructions
- Request and install twwistPDB
(if you have not already done so).
https://weiningerworks.com/contact.html - Download PDB structure files from rcsb.org (if you have not already done so):
1W1X, 2KS9
- Download PDB structure files from weiningerworks.com:
WSUBP.pdb (substance P with side chains reoriented to match the N6 neuraminidase)
- Run twwistPDB to orient WSUBP.pdb to 1W1X.pdb.
twwistPDB will output a single reoriented pdb file, WSUBPontoN6.pdb.
cp 1W1X.pdb N6.pdb
twwistPDB -o WSUBPontoN6.pdb SUBP.pdb 5869 5895 5909 1W1X.pdb 8715 8726 8733
- The new pdb file, WSUBPontoN6.pdb, shares the same orientation as 1W1X.pdb (“N6.pdb”).
Open WSUBPontoN6.pdb and N6.pdb at the same time in a molecular visualization program. Note that the twwistPDB command superimposed common substance P and N6 neuraminidase residues.
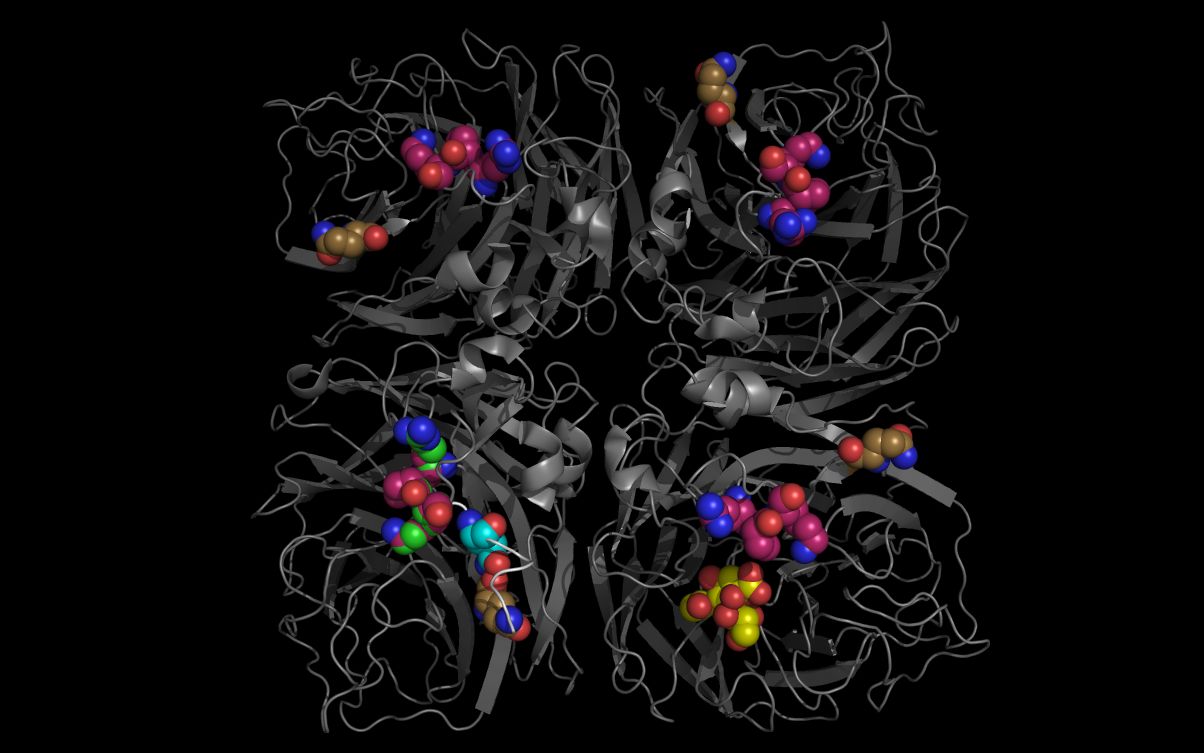
The picture above shows:- Substance P cartoon colored white
- N6 neuraminidase tetramer cartoon colored grey
- Sialic acid carbon spheres colored yellow
- Substance P residue R365, P366, K367 carbon spheres colored green
- Substance P residue Q369 residue carbon spheres colored cyan
- N6 neuraminidase tetramer residue R438, P439, K440 carbon spheres colored magenta
- N6 neuraminidase tetramer residue Q407 residue carbon spheres colored gold
The flexible R365-K367 substance P side chains have been repositioned onto the R428-K440 N6 neuraminidase side chains. The repositioning of the loops containing these residues would bring the Q369 and Q407 into spatial overlap.
The alignment of putative cell entry domain residues in substance P (residues R365, P366, K367, Q369) and on the N6 neuraminidase residues on the surface of the N6-containing virions (1W1X residues R438, P439, K440, and Q407) have not been optimized as the main and side chain atoms of N6 have not been moved to align the Q369 residue from substance P and the Q407 residue from N6 neuraminidase.