Weininger Works™ Open Access Method: Molecular Locks
Overview
Configuration, Sensitivity, and Specificity
Circuits
Open Access
(WEB PAGE PDF)
Molecular Lock Overview
Molecular Locks:
- were invented, patented, and developed by Arthur Weininger and Susan Weininger;
- are protein assemblies that cooperatively lock onto specific target nucleic acids (DNA or RNA); and
- form the basis of our Molecular Lock Pathogen Shield (MOLOPS) system.
MOLOPS nucleic acid detection components:
- can be cost-effectively made in bacteria;
- can be easily tested and evaluated in gel retardation assays;
- are highly stable to environmental factors (e.g. temperature);
- can be used on crude lysates; and
- are able to be implemented in many formats (e.g. dipstick or electronic).
MOLOPS nucleic acid silencing application components:
- can be transferred to cells as proteins or encoded in deliverable nucleic acids; and
- most often are designed to target:
- replication control sequences that are upstream of coding regions (e.g. HIV LTR); or
- promoters that control disease-causing or disease-permitting genes
(e.g. pathogenic Burkholderia pseudomalei toxin protein and drug resistance protein promoters).
We believe that it is critical that a global MOLOPS surveillance system be set up to identify, characterize, and inactivate pathogenic biota. The detection of pathogenic biota and the silencing of pathogenic promoters and control regions provide the basis for a broad pathogen shield.
Molecular Lock Configuration, Sensitivity, and Specificity
Molecular Locks are molecular assemblies that are engineered to bind specifically to nucleic acid targets. Target specificity is obtained even if the target is comprised of commonly occurring sequences. Molecular Lock components can be used to protect or isolate the target. In tests, non-target nucleic acids can be eliminated without degrading or reducing the target nucleic acids.
Configuration
Molecular Lock oligomerization domains are selected so that nucleic acid binding domains cooperatively bind to the target in a specific geometry.

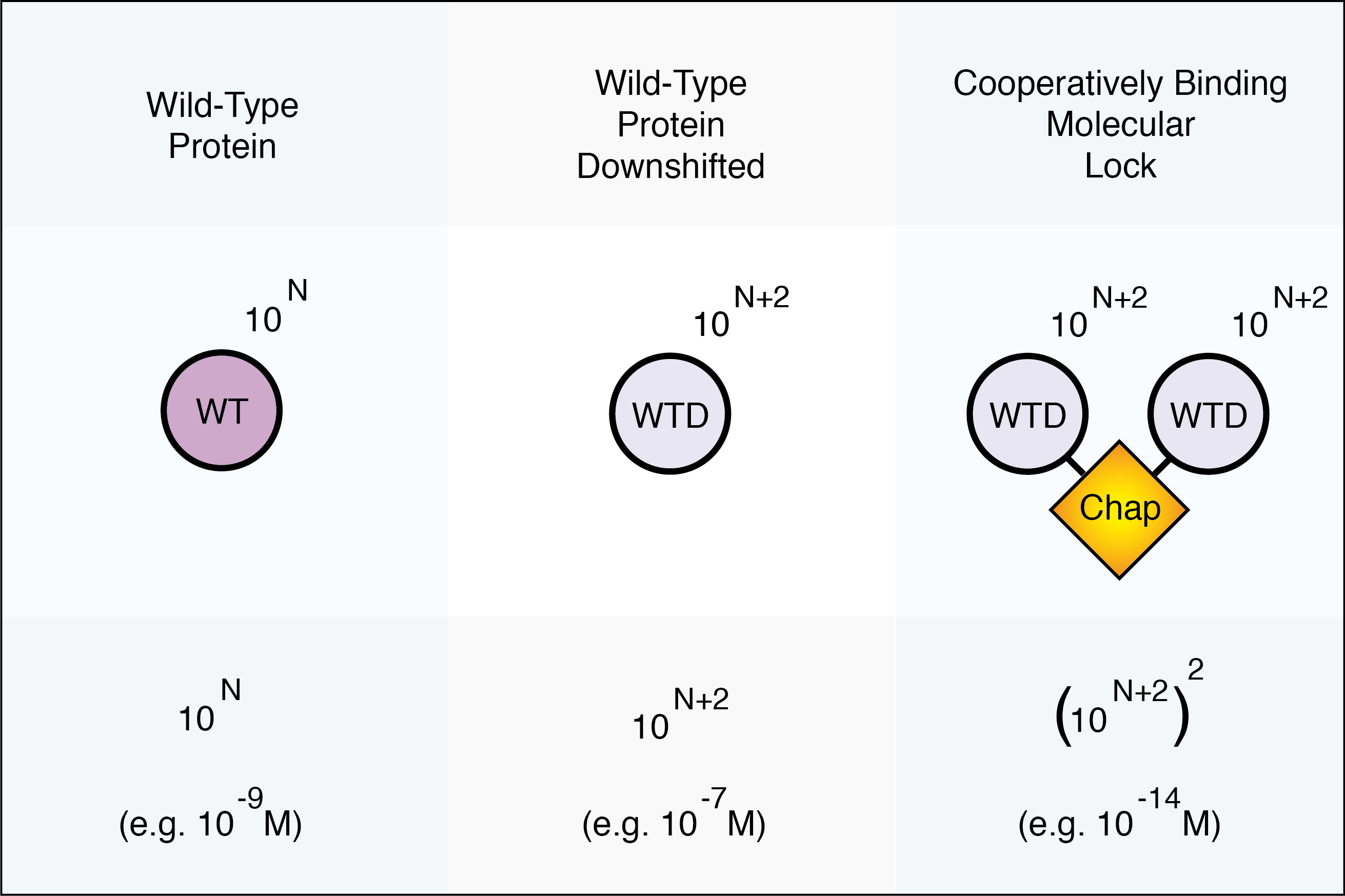
Sensitivity
The binding affinity of a molecular assembly that binds cooperatively with two binding domains is on the order of the square of the binding affinity of a molecule binding with only one of the domains. Downshifting the binding affinity of each of the Molecular Lock components allows the assembled block to bind tightly to the target even if each subcomponent would be weakly bound individually. This is an important feature both in vitro and in vivo.
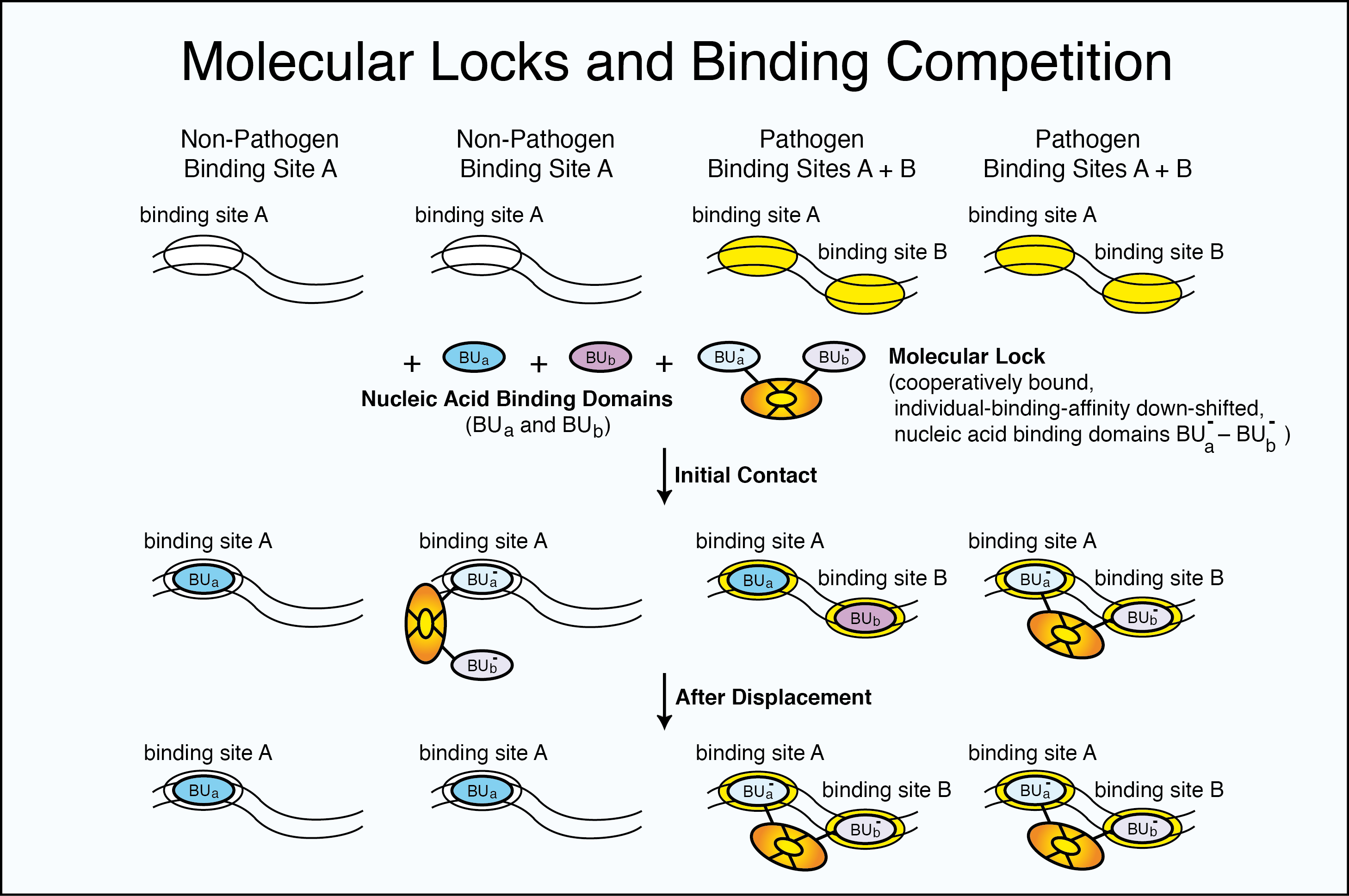
Specificity
Molecular Locks formed from more than one binding component can select specific nucleic acid targets
—even when a portion of the target is found elsewhere in a non-target sequence.
Molecular Lock Circuits
Molecular Locks can also be used to characterize molecular circuits. Characterized molecular circuits can be used to design specific or broad Molecular Locks.
Download video or right mouse down (on video) to “Save As...”
Susan Weininger
EE 380 Computer Systems Colloquium
Stanford University
14 October 2009
Molecular Lock Method Open Access
Molecular Lock method patents have been broadly issued to Arthur Weininger and Susan Weininger. These patents have now expired. The Molecular Lock method described in the expired patents is now an Open Access Method. Note that individual Molecular Locks may still be individually protected as composition patents.
Open Access Methods (Definition and Disclaimers)
“Open Access Methods” are specific methods upon which Arthur Weininger and Susan Weininger make no proprietary claim.
Open Access Methods listed by Arthur Weininger and Susan Weininger are provided “as is” and without warranty of any kind.
No oral or written information or advice given by Arthur Weininger and/or Susan Weininger shall create any warranty with regards to Open Access Methods.
Open Access Methods may not be: useful, safe, or non-infringing of third party rights.
Arthur Weininger and Susan Weininger do not agree to any indemnification for anything related to Open Access Methods including, but not limited to,
any use of Open Access Methods, any legal actions directly or indirectly associated with Open Access Methods, or any losses or damages directly or indirectly associated with Open Access Methods.
In no event shall the total liability by Arthur Weininger and Susan Weininger for any and all damages related to Open Access Methods exceed zero.
Any use of Open Access Methods is at your sole and entire risk.