combinePDB
Overview
combinePDB Help Output
combinePDB Tutorial
(WEB PAGE PDF)
combinePDB Overview
combinePDB is a program that combines two PDB files into one PDB file.
combinePDB Help Output (“combinePDB -h” output)
NAME
combinePDB (version 1.1.1) -- combines two PDB files into one PDB file
SYNOPSIS
combinePDB [options]
CHARACTER OPTION___KEYWORD OPTION_______DESCRIPTION__________________________DEFAULT__________
-i <filename> .. --input1=<filename> .. first input PDB file ............... stdin
-j <filename> .. --input2=<filename> .. second input PDB file .............. none (required)
-o <filename> .. --output=<filename> .. output PDB file .................... stdout
-a <model#> .... --model1=<model#> .... MODEL # of first input PDB file ... first model
-b <model#> .... --model2=<model#> .... MODEL # of second input PDB file ... first model
-c <chainids> .. --chainids=<str> ..... user replacement chain IDs ......... none
-m <filename> .. --map=<filename> ..... generate file of atom changes ...... no map
-n ............. --noidchange ......... do not change input PDB chain IDs .. chain IDs changed
-p ............. --nopositionchange ... do not change position of records .. positions changed
-x ............. --noresiduechange .... do not change residue numbers ...... residues changed
-h ............. --help ............... prints help (Enter 'combinePDB -h' for help.)
<NO OPTIONS> .......................... shorter option synopsis (Enter 'combinePDB'.)
--license ............ prints license terms for combinePDB.
DESCRIPTION
combinePDB combines two PDB files into one PDB file.
combinePDB reads two existing input PDB files and writes a new output PDB file.
The output PDB file will contain all specified ATOM, HETATM, TER, and CONECT records
from both input files. CONECT records will be updated to reflect new indexing.
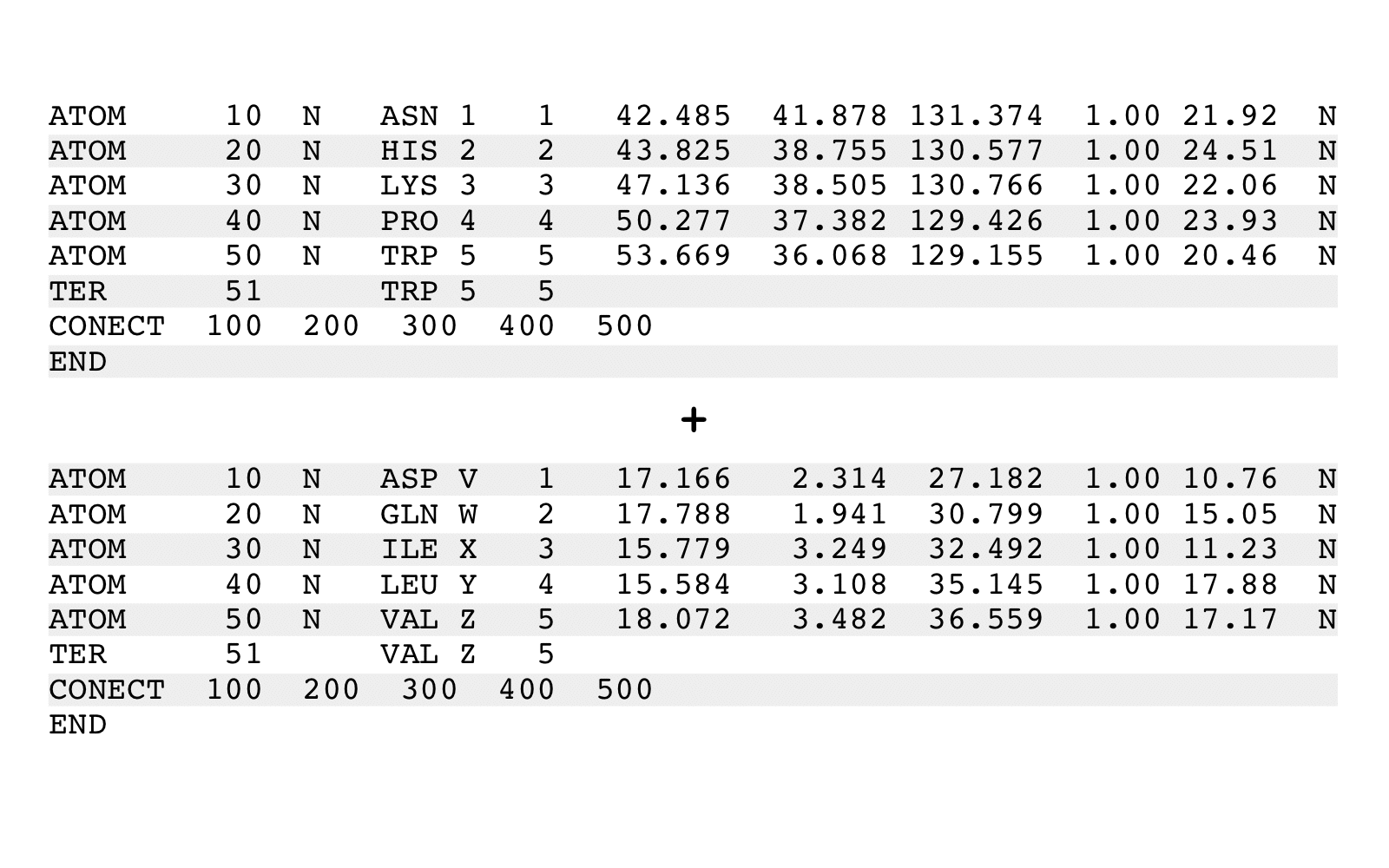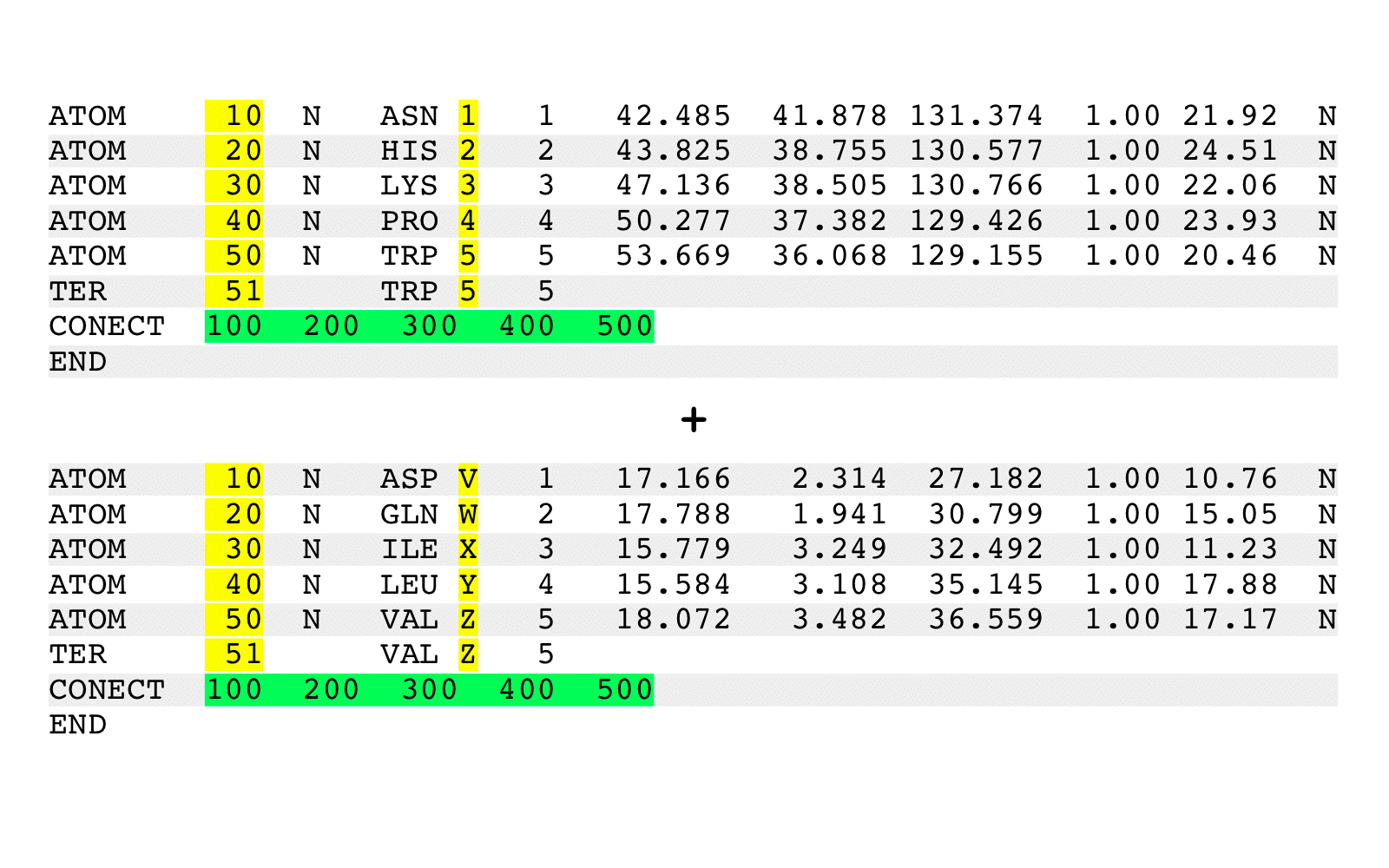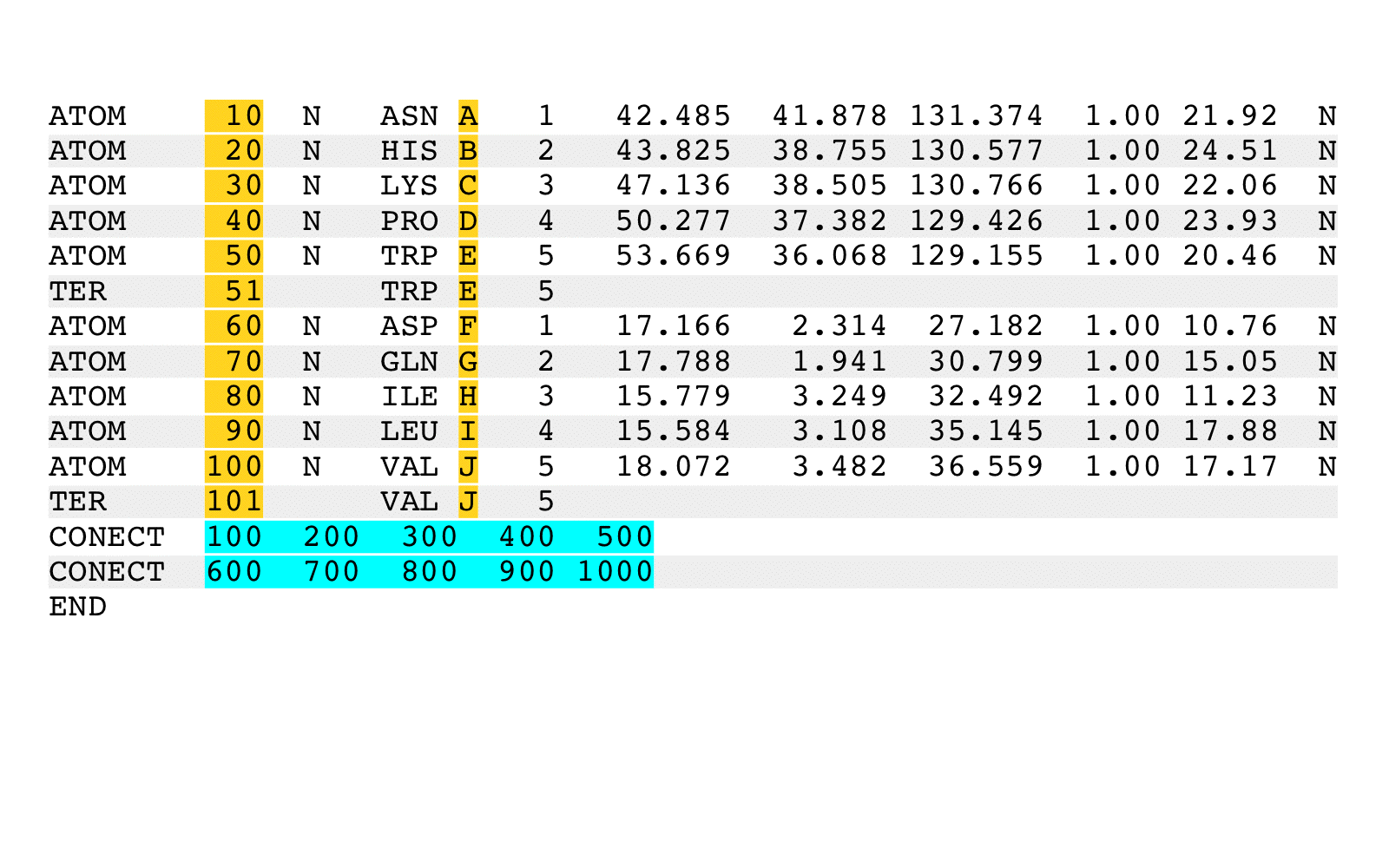
By default, chain identifiers and residue serial numbers will be assigned unique
values, and chains will be written out sequentially by new alphanumeric chain order.
Optionally, record order, chain identifiers, and residue serial numbers may be preserved.
Input files are expected to be in PDB format. Option '--input1=' ('-i') specifies
the first input filename. If option '--input1=' ('-i') is not present, input will
be read from stdin. Option '--input2=' ('-j') specifies the second input filename.
Option '--model1=' ('-a') may be used to specify the MODEL number of the first input
PDB file to process. Option '--model2=' ('-b') may be used to specify the MODEL
number of the second input PDB file to process. MODEL option values of '1' or more
will match the first encountered MODEL having the specified MODEL number. A MODEL
option value of '0' will match the first (implicitly or specifically labelled) MODEL.
By default (i.e., when no '--model#=', '-a', or '-b' is used), the first MODEL
(perhaps implicit if no MODEL records exist) from each input file is processed;
this is equivalent to '--model1=0' and '--model2=0' ('-a 0' and '-b 0').
Option '--output=' ('-o') specifies the output filename. If option '--output=' ('-o')
is not present, output will be written to stdout.
New "REMARK 250" lines will be added to the output PDB specifying execution details.
By default, chain identifiers will be altered as required to produce an output file
with unique chain identifiers. Chain identifiers are filled in the order of the
94 listed characters below (i.e., the first listed are the first used):
'A' to 'Z'
'0' '1' '2' '3' '4' '5' '6' '7' '8' '9'
'a' to 'z'
'!' '"' '#' '$' '%' '&' ''' '(' ')' '*' '+' ',' '-' '.' '/' ':'
';' '<' '=' '>' '?' '@' '[' '\' ']' '^' '_' '`' '{' '|' '}' '~'
Option '--chainids=' ('-c') specifies a string of user supplied replacement chain
identifiers, allowing chains of the combined output PDB file to be arbitrarily labeled.
The user supplied replacement chain identifier string can be any size, can be set with
any non-NULL character, and can contain multiple occurrences of the same character.
If the user supplied replacement chain identifier string starts and ends with either
a single quote (''') or a double quote ('"'), then these quotes will be treated as
string delimiters and not as chain indentifiers. When quotes are intended to be used
as chain indentifiers, then the quote characters must: (i) be listed inside a quoted
string and (ii) be preceded with a single backslash character ('\').
[The wwPDB states that "The use of punctuation characters in the place of alphanumeric
characters is discouraged." However, it is sometimes useful for temporary PDB files.]
Non-contiguous input PDB file records that have identical chain indentifiers will
be output with the associated output records having identical chain identifiers.
If there are more chains in the combined output PDB file than there are replacement
chain identifiers (either by default or user-supplied) then a space (' ') will be
assigned as a chain identifier for all chains after the last replacement chain
identifier has been assigned. If this occurs then a warning will be given (written
to stderr). The space character is not one of the default replacement characters.
[For more flexibility in setting chain identifiers, use 'chainidPDB' after using
combinePDB. 'chainidPDB' is a PDB filter for changing one or all chain identifiers.
'chainidPDB' is a unix command line program from Weininger Works
(www.weiningerworks.com).]
Option '--noidchange' ('-n') may be used to specify that input chain identifiers are
to remain unchanged.
By default, combinePDB fixes chains that have residues with duplicate residue sequence numbers.
Option '--noresiduechange' ('-x') may be used to specify that input residue numbers are to
remain unchanged; a warning will be generated when ambiguous residue numbers are output.
By default, atoms will be written out sequentially by new alphanumeric chain order. The
new chain order will consist of any existing or specified chains in the following order:
'A' - 'Z'
'0' - '9'
'a' - 'z'
' ' (space)
< all other ASCII values from 1 to 127, inclusive >
When the '--nopositionchange' ('-p') option is used, the output PDB file will contain all
specified ATOM, HETATM, TER, and CONECT records from both input files in the following order:
ATOM, HETATM, and TER records from first input PDB file
ATOM, HETATM, and TER records from second input PDB file
CONECT records from first input PDB file
CONECT records from second input PDB file
The option '--map=' ('-m') may be used to generate a text file listing atom changes.
No map file is generated by default.
Any errors and warnings will be written to stderr.
EXAMPLE
The following will combine in1.pdb and in2.pdb to out.pdb.
With keyword options:
combinePDB --input1=in1.pdb --input2=in2.pdb --output=out.pdb
With character options:
combinePDB -i in1.pdb -j in2.pdb -o out.pdb
The following will combine in1.pdb and in2.pdb to out.pdb, and
specifies that the chains in out.pdb will be labeled with chain
identifiers taken in order from the string "13579acegikmoqsuwy".
With keyword options:
combinePDB --input1=in1.pdb --input2=in2.pdb --output=out.pdb \
--chainids="13579acegikmoqsuwy"
With character options:
combinePDB -i in1.pdb -j in2.pdb -o out.pdb -c "13579acegikmoqsuwy"
LICENSE INFORMATION
combinePDB is a software program from Arthur Weininger (weiningerworks.com).
combinePDB is subject to a license; use the keyword option '--license' in order to view
the license terms. Your use of this software contitutes an agreement to the license terms.
Do not use this software if you do not agree to the license terms.
combinePDB Tutorial
The Picornavirus Monograph Superposition Shell Script gives examples of using combinePDB.