Weininger Works™ Open Access Molecule: FLU-LOCK
Overview
Emergent Influenza
Structural Basis
Open Access
(WEB PAGE PDF)
FLU-LOCK Overview
We (Arthur Weininger and Susan Weininger) continue to be concerned about unusual emergent animal and human influenza viruses. We have published a structural analysis of a set of highly unusual influenza viruses, H18N11 and H17N10. We aligned the sequences (in the absence of sequence homology) and structures of the sialidases. Among other unusual features, we found toxin domains on the N11 and N10 proteins. We determined the structural bases in the N11 and N10 proteins for the alarming absence of sialidase activity and for the alarming absence of anti-viral binding group configurations where the normal sialic acid/drug binding site should be. We screened the influenza capsids and isolated non-active site regions (outside of the normally conserved sialic acid binding site) that could be used to bind anti-virals. We determined that small molecules in a set of nucleoprotein crystal structures presented better binding profiles if reoriented (rotated by ~90 degrees). We designed FLU-LOCK to bind tightly and specifically to one of the conserved sites in the nucleoprotein complex.
Emergent Influenza
New influenza viruses emerge regularly. For example, the H17N10 and H18N11 viruses isolated from bats in Guatemala (H17N10) and Peru (H18N11) are highly divergent. The N10 and N11 neuraminidase proteins have no functional sialidase site and have no sequence homology to other neuraminidases.
We studied the structural and sequence invariants and divergences of neuraminidases.
dx.doi.org/10.1371/journal.pone.0117499
(16 MB PDF – LOCAL COPY) (3 MB PDF – LOCAL COPY)
We aligned the structures and sequences of N10 protein (N10P), N11 protein (N11P), an influenza A neuraminidase (N6N), an influenza B neuraminidase (IBN), and a bacterial neuraminidase. This allowed us to compare the active and inactive sites of these proteins and to further evaluate the compatibility of the inactive sites of the N10 and N11 neuraminidases with the structures of drug compounds that target the normally conserved sialic acid site. The following image shows aligned N6N, N10P, N11P, and IBN sialidases, both individually and in a combined superposition:
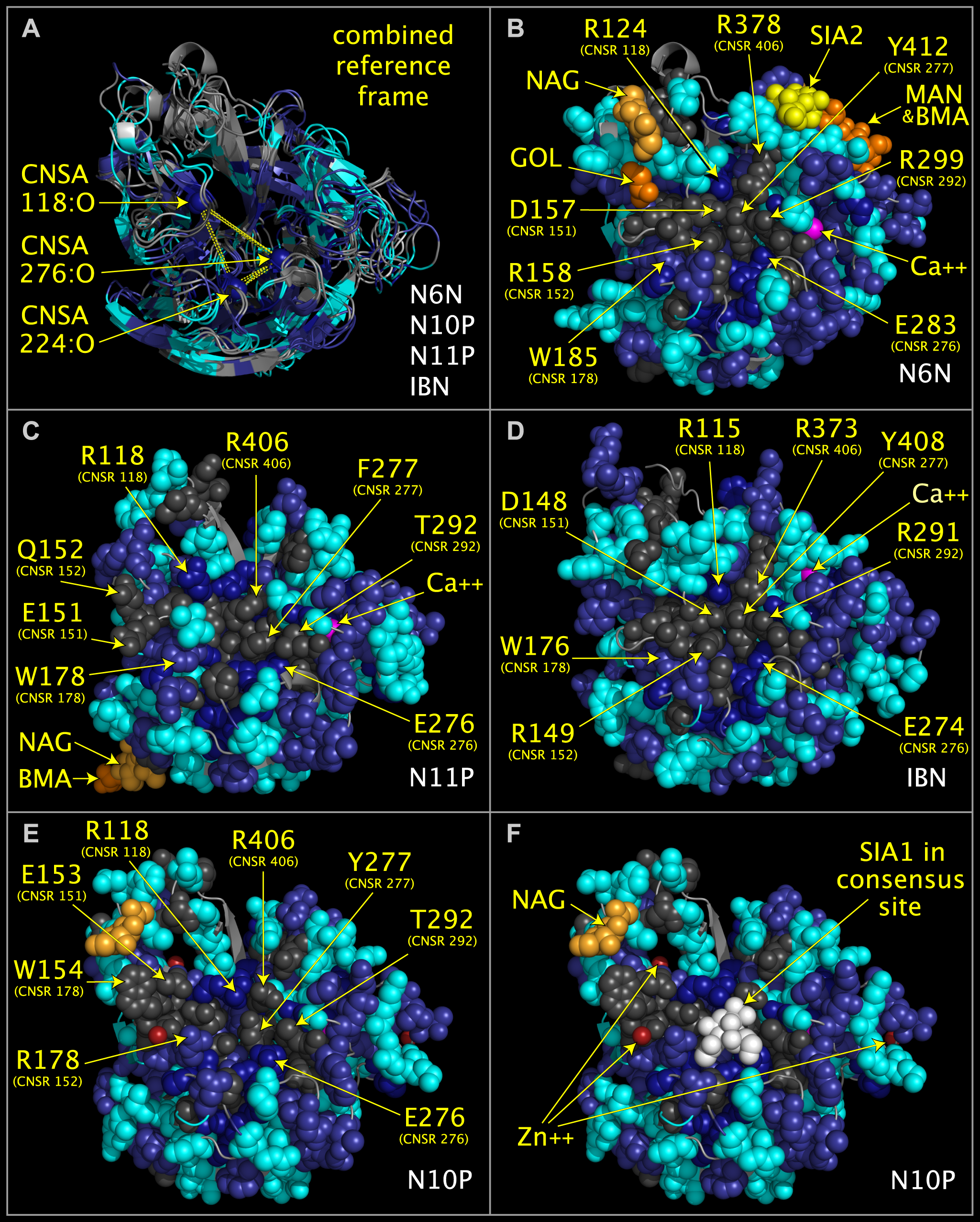
We found similarities between structures in the N10 and N11 virus capsid proteins and structures in non-neuraminidase proteins. Among other findings, we found that the N10 and N11 capsid proteins contain toxin-like domains. In N10 protein, we identified staphylococcal enterotoxin I-like domains. In N11 protein, we identified hepatitis E E2S-like domains, SARS spike protein-like domains, and toxin components shared by alpha-bungarotoxin, staphylococcal enterotoxin I, anthrax lethal factor, clostridium botulinum neurotoxin, and clostridium tetanus toxin. The following image shows the N11 protein, with its toxin-like domains highlighted, next to toxins containing structurally similar residue clusters.
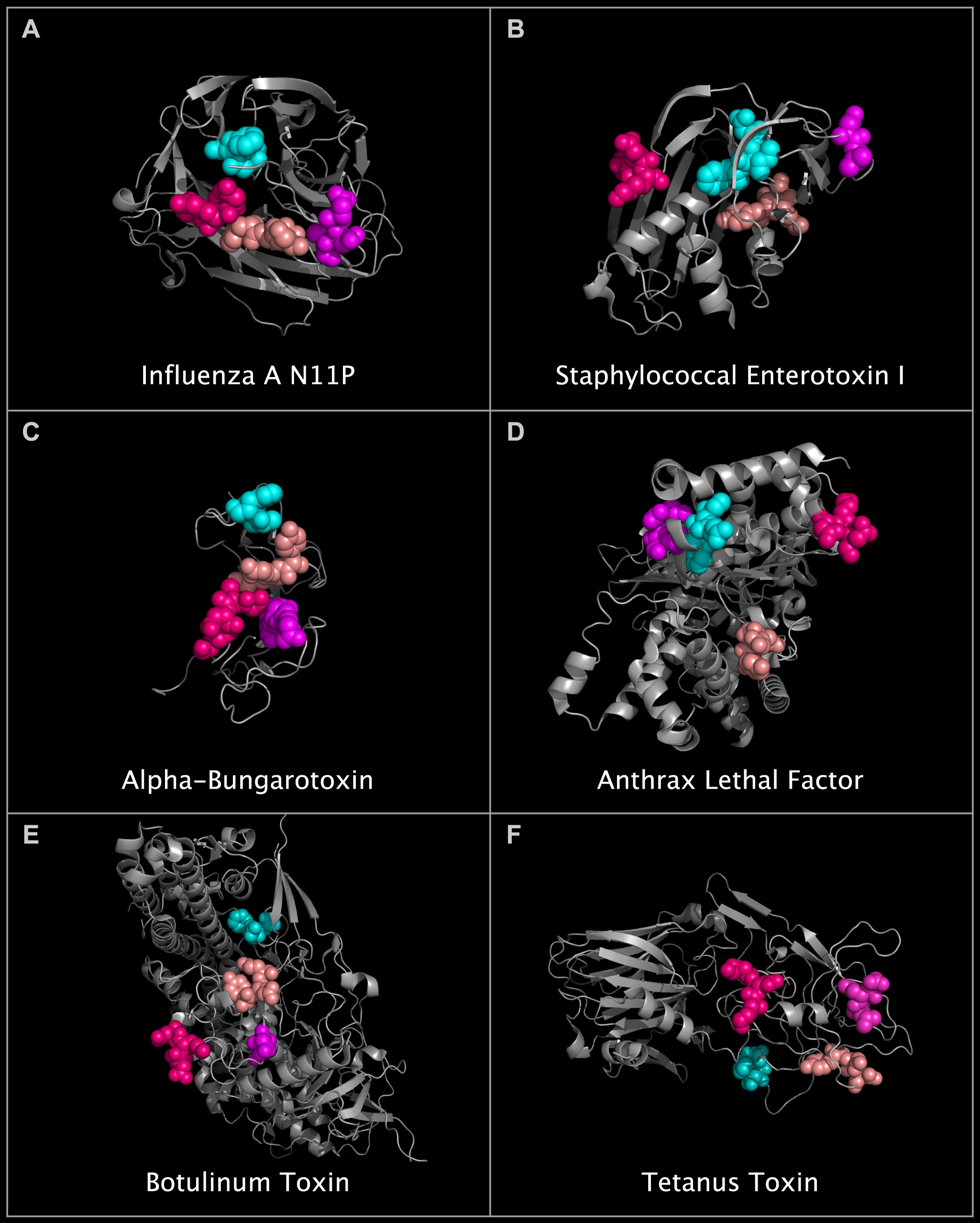
We concluded that the absence of demonstrated neuraminidase activity with the presence of new cell entry domain components in N10 and N11 proteins suggests that N10 and N11 protein-containing viruses may enter cells without a functioning sialidase, i.e., by binding to alternative receptors such as ACE2, acetylcholine, and MHC II receptors on an expanded receptive cell population, including cells such as neurons and T-cells.
FLU-LOCK Structural Basis
While screening protein surfaces presented by influenza virions, we noticed potential problems with structures containing a class of small molecules bound to nucleoproteins. We examined the nucleoprotein superstructure and found an alternative positioning (rotation in place) of small molecules bound to influenza nucleoproteins. This repositioning appears to resolve all chemical and structural issues that we noticed.

The rotation of the bound molecules provided a better physical and chemical fit and suggests an antiviral molecule, FLU-LOCK. Because FLU-LOCK is a dimer, it is expected to have much tighter affinity for the nucleoprotein site than single components.
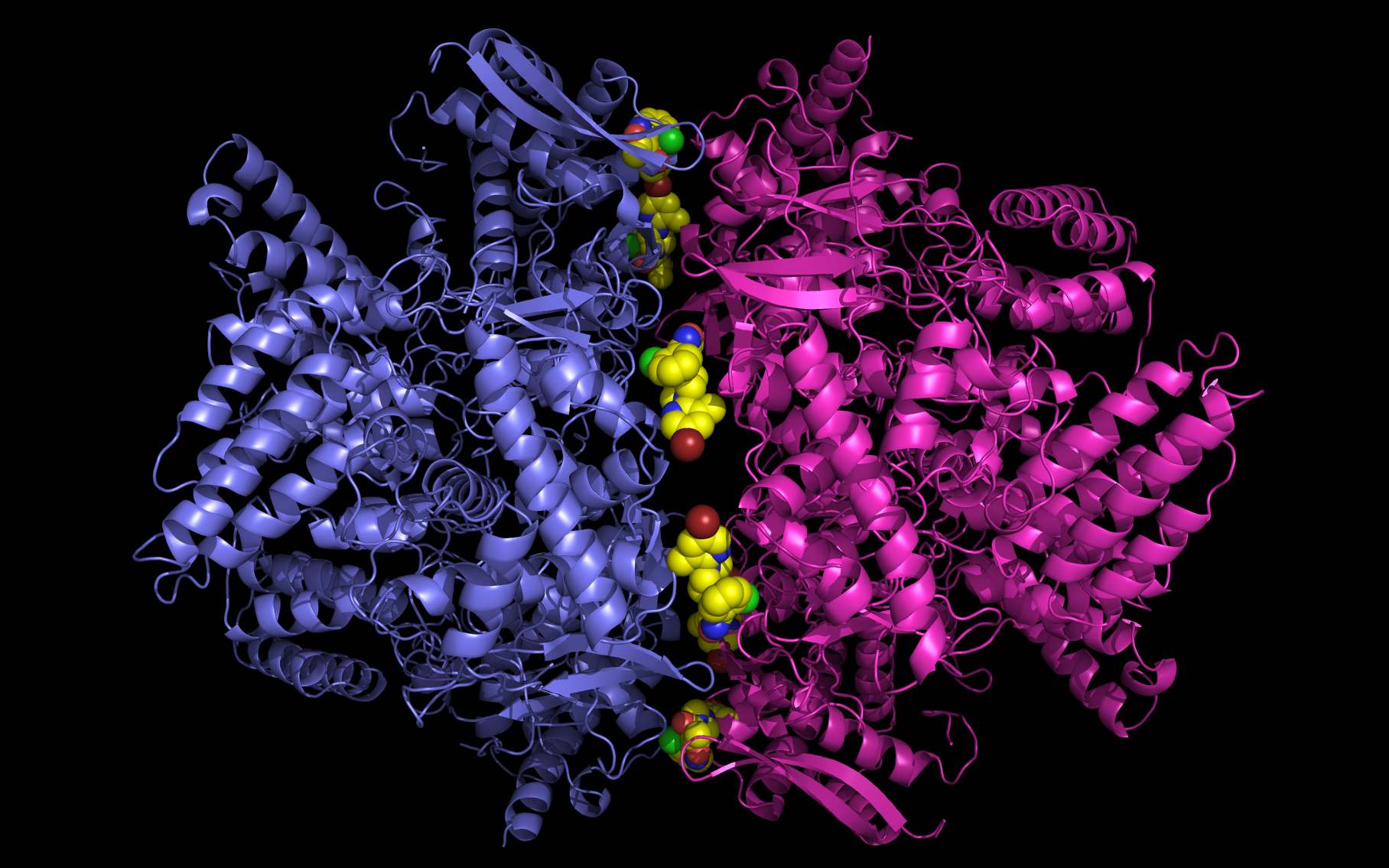
Weininger Works Technical Notes (2013) Sept 10;4:1-22 (HTML) (PDF)
FLU-LOCK Open Access Molecule
FLU-LOCK is an Open Access Molecule.
Open Access Molecules (Definition and Disclaimers)
“Open Access Molecules” are specific molecules upon which Arthur Weininger and Susan Weininger make no proprietary claim.
Open Access Molecules listed by Arthur Weininger and Susan Weininger are provided “as is” and without warranty of any kind.
No oral or written information or advice given by Arthur Weininger and/or Susan Weininger shall create any warranty with regards to Open Access Molecules.
Open Access Molecules may not be: useful, safe, or non-infringing of third party rights.
Arthur Weininger and Susan Weininger do not agree to any indemnification for anything related to Open Access Molecules including, but not limited to,
any use of Open Access Molecules, any legal actions directly or indirectly associated with Open Access Molecules, or any losses or damages directly or indirectly associated with Open Access Molecules.
In no event shall the total liability by Arthur Weininger and Susan Weininger for any and all damages related to Open Access Molecules exceed zero.
Any use of Open Access Molecules is at your sole and entire risk.