dist
Overview
dist Help Output
dist Tutorial
(WEB PAGE PDF)
dist Overview
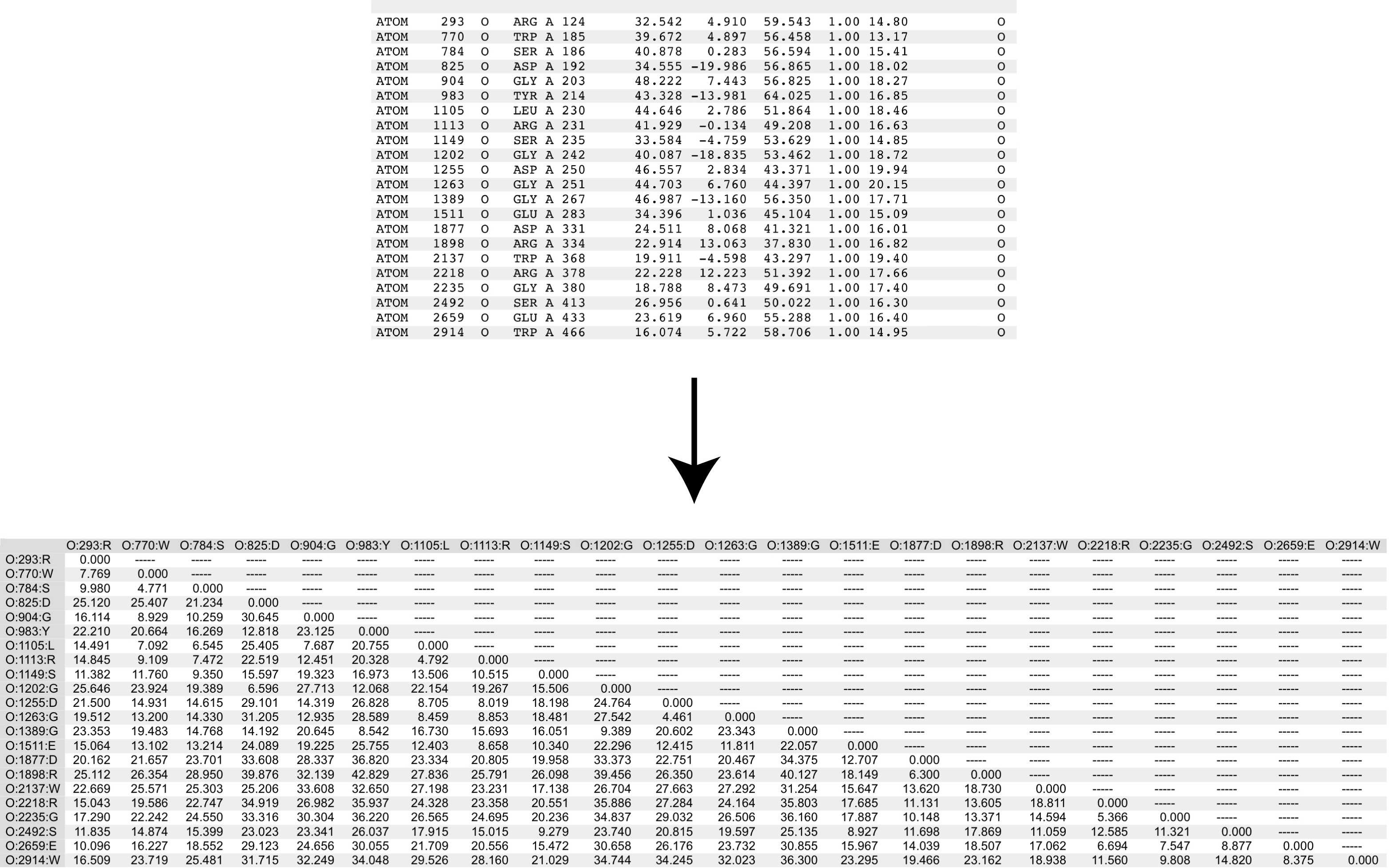
dist creates a distance matrix from a points list, generating a lower triangle matrix with distances between all the points, a zero-valued diagonal, and an upper triangle matrix with hyphens or zeroes (e.g. '---' or '0.00'). There are options for specifying upper triangle contents, handling input and output labels, and output format. dist can optionally parse a PDB file, and has options for specifying user defined output atom labels.
dist Help Output (“dist -h” output)
NAME
dist (version 1.0.2) -- creates distance matrix from points list
SYNOPSIS
dist [options]
CHARACTER OPTION_____KEYWORD OPTION________DESCRIPTION____________________________DEFAULT____
Options for input:
-i <filename> .... --input=<filename> .... input points list filename ........... stdin
-n ............... --no_input_labels ..... input points list has no labels ...... has labels
Options for output:
-o <filename> .... --output=<filename> ... output distance matrix filename ...... stdout
-f #.# .......... --format=#.# .......... real number format (width.precision) . 5.3
-s # ............. --spaces=# ............ separate fields with # spaces ........ 0 (tabs used)
-t ............... --truncate_labels ..... action to fit col width to labels .... no truncation
-x ............... --no_output_labels .... do not output labels ................. output labels
-z ............... --upper_zeroes ........ fill upper matrix with zeroes ........ '---'
Options for PDB input and atom label output:
-a [<labelfmt>] .. --atom_labels ......... input points list is PDB file ........ not PDB file
--atom_labels=<labelfmt>
<labelfmt> characters specify atom label content ............................. '@:#:*'
@ _______________ atom name __________________ <= 3 characters
# _______________ atom serial number _________ <= 5 digits
& _______________ residue name _______________ 3 characters
* _______________ residue name _______________ 1 character
? _______________ residue sequence # _________ <= 4 digits
% _______________ chain identifier ___________ 1 character
Other options:
-d ............... --info ................ outputs processing info to stderr .... no info
-h ............... --help ................ prints help (Enter 'dist -h' for help.)
--license ............. prints license terms for dist.
DESCRIPTION
dist creates a distance matrix from a points list, generating a lower triangle matrix
with distances between all the points, a zero-valued diagonal, and an upper triangle
matrix with either hyphens or zeroes (e.g. '---' or '0.00').
By default, input comes from stdin and output goes to stdout.
Option '--input=' ('-i') allows command line specification of the input file.
Option '--output=' ('-o') allows command line specification of the output file.
Errors and warnings go to stderr.
The points list should consist of points on separate lines as "[label] x y z", i.e.
an optional label followed by 3 real numbers. Labels and real numbers must be separated
by white space (i.e., spaces or tabs). Labels are assumed to be present in the input
points list file by default. If there are no labels in the input points list file, then
the option '--no_input_labels' ('-n') must be used and no input labels will be parsed.
Labels are output as the topmost row and the leftmost column by default.
The option '--no_output_labels' ('-x') can be used to suppress all labels from the output.
In the case where option '--no_output_labels' ('-x') is not used (and labels are to be
output by default) and option '--no_input_labels' ('-n') is used at the same time (and no
labels are read in from input), then output labels will be generated as 'pt#'.
Distance values are output as real numbers. The option '--format=' ('-f') must be followed
by an option value specifying the output format as 'mmm.ddd', where 'mmm' is the minimum
field width for the entire real number, and where 'ddd' is the precision. The default
format is '5.3', where the minimum field width for the entire number is 5 characters,
and there are 3 digits following the decimal point.
By default, tabs delimit output columns and spaces are not output. The option '--spaces='
('-s') specifies that spaces will be output to delimit column values instead of tabs; the
option '--spaces=' ('-s') must be followed by an integer option value specifying the number
of spaces to be used.
If spaces are specified to be used instead of tabs, column width can either be expanded
to the size of each label associated with that individual column or labels can be truncated
to a common column size. The default behaviour is to expand any specific column width to
fit its label. The option '--truncate_labels' ('-t') truncates, if necessary, the labels'
to fit a common column size.
By default, hypens (i.e., '---') are output for the values of the upper triangle matrix.
Option '--upper_zeroes' ('-z') changes these output values to zeroes (e.g., '0.00').
Option '--atom_labels=' ('-a') allows parsing of an atom list provided in 'PDB' file format.
An optional accompanying option value specifies the label format, i.e., the content of the
labels. Six reserved characters are used to specify substitution with specific fields from
'ATOM' or 'HETATM' pdb lines:
Char Substitution Size
---- ------------------------ -------------------
@ ... atom name ................. <= 3 char output
# ... atom serial number ........ <= 5 digits output
% ... chain identifier .......... 1 char output
& ... residue name .............. 3 char output
* ... residue name .............. 1 char output
? ... residue sequence number ... <= 4 digits output
All other characters specified in the option value will be directly used in each label.
Labels will be written out in the order of the position of all characters in the option
value. When option '--atom_labels=' ('-a') is used without an option value, the default
label format specification is '@:#:*', and would produce labels that look similar to
'NH1:1171:R'. Note: If you use option '--atom_labels=' ('-a') without an accompanying
option value, make sure to put '--atom_labels=' ('-a') at the end of the command. Reserved
characters can be directly used without being substituted by: (1) adding single quotes to
front and back of the label format option value, and also (2) using a backslash in front
of the reserved characters that are intended to be directly used without substitution;
e.g., "-a 'ATOM\##'" will produce labels that look similar to 'ATOM#1171'. Note: Always
use single quotes on front and back of the label format option value; this will keep the
reserved characters from being used as unix shell directives.
Option '--info' ('-d') is used to output, to stderr, processing information: number of
points read, the list of points, and completion statement. No processing information
is output by default.
Option '--help' ('-h') prints this help.
EXAMPLES
The following command line would read input, including labels, from the file 'points_list'
and write a new file, named 'distance_matrix', with the contents of a labelled, tab-delimited
distance matrix.
With keyword options:
dist --input=points_list --output=distance_matrix
With character options:
dist -i points_list -o distance_matrix
The following command line would read input, including labels, from the file 'points_list'
and write a new file, named 'distance_matrix', with the contents of a labelled, space-delimited
distance matrix.
With keyword options:
dist --input=points_list --output=distance_matrix --spaces=2
With character options:
dist -i points_list -o distance_matrix -s 2
The following command line would read input from the file 'points_list' (a file not containing
labels) and write a new file, named 'distance_matrix', with the contents of a space-delimited
distance matrix with no labels, and zero values for the upper triangle matrix.
With keyword options:
dist --input=points_list --output=distance_matrix --spaces=2 \
--no_input_labels --no_output_labels --upper_zeroes
With character options:
dist -i points_list -o distance_matrix -s 2 -n -x -z
The following command line would read input from the file 'atoms.pdb' and write a new file,
named 'distance_matrix', with the contents of a tab-delimited distance matrix with labels
that look similar to 'NH1:1171:R' (by default).
With keyword options:
dist --input=atoms.pdb --output=distance_matrix --atom_labels
With character options:
dist -i atoms.pdb -o distance_matrix -a
LICENSE INFORMATION
dist is a software program from Arthur Weininger (www.weiningerworks.com).
dist is subject to a license; use the keyword option '--license' in order to view
the license terms. Your use of this software contitutes an agreement to the
license terms. Do not use this software if you do not agree to the license terms.
dist Tutorial
dist and deviation Tutorial Page gives examples of using dist.