Mouse over the small images above to change larger image and associated description. (Alternatively, read static PDF.)
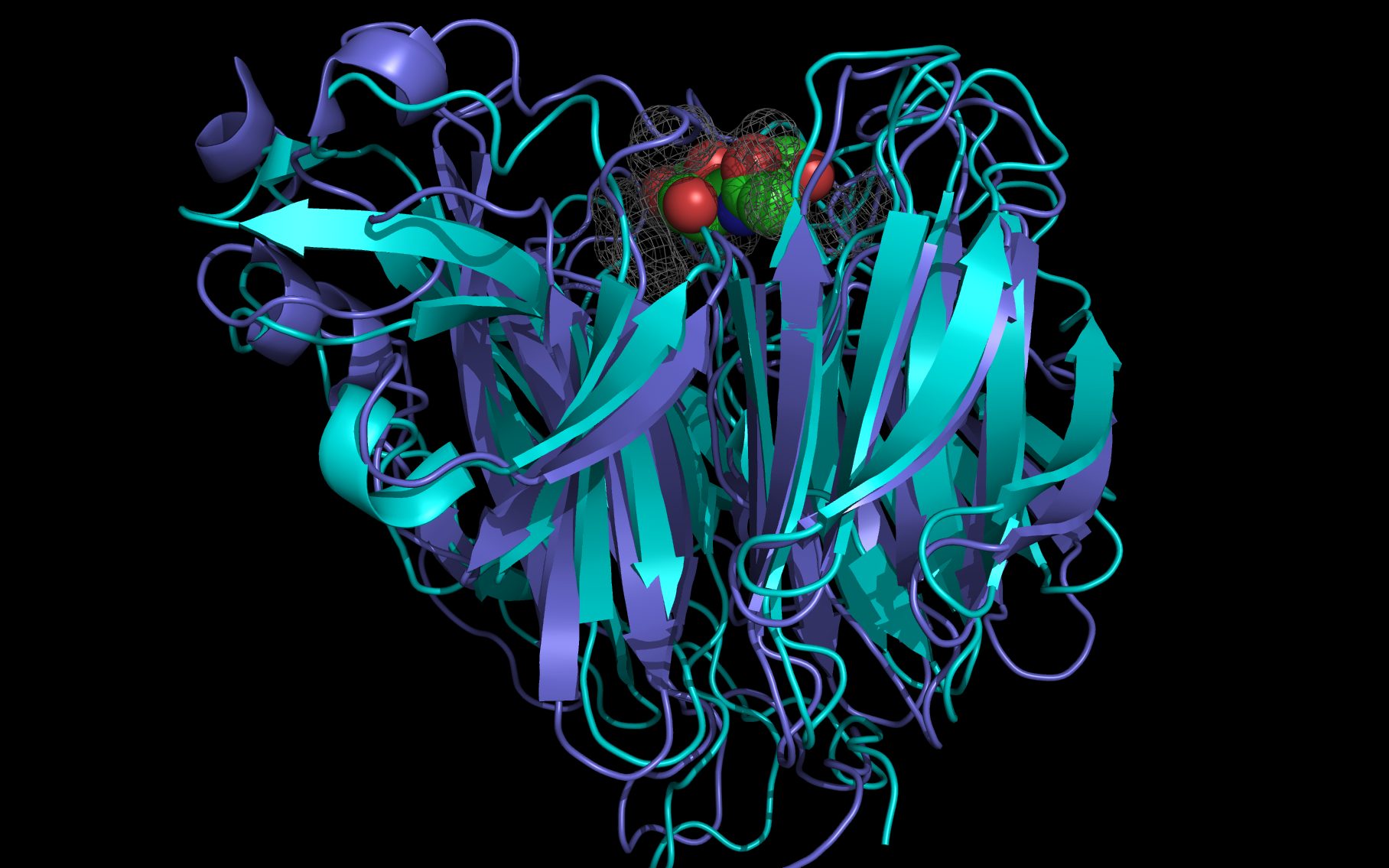
The bat influenza virus N10 neuraminidase has 20 to 27 percent sequence identity with other influenza virus sialidases. wwavePDB analysis suggests that the bat influenza virus N10 neuraminidase is a viral enzyme that has maintained its sialic acid binding site without maintaining sequence homology with other influenza virus sialidases. The following pictures and description document the finding of key catalytic residues and the building of a putative sialic acid binding site using wwavePDB and the Weininger Works' program twwistPDB.
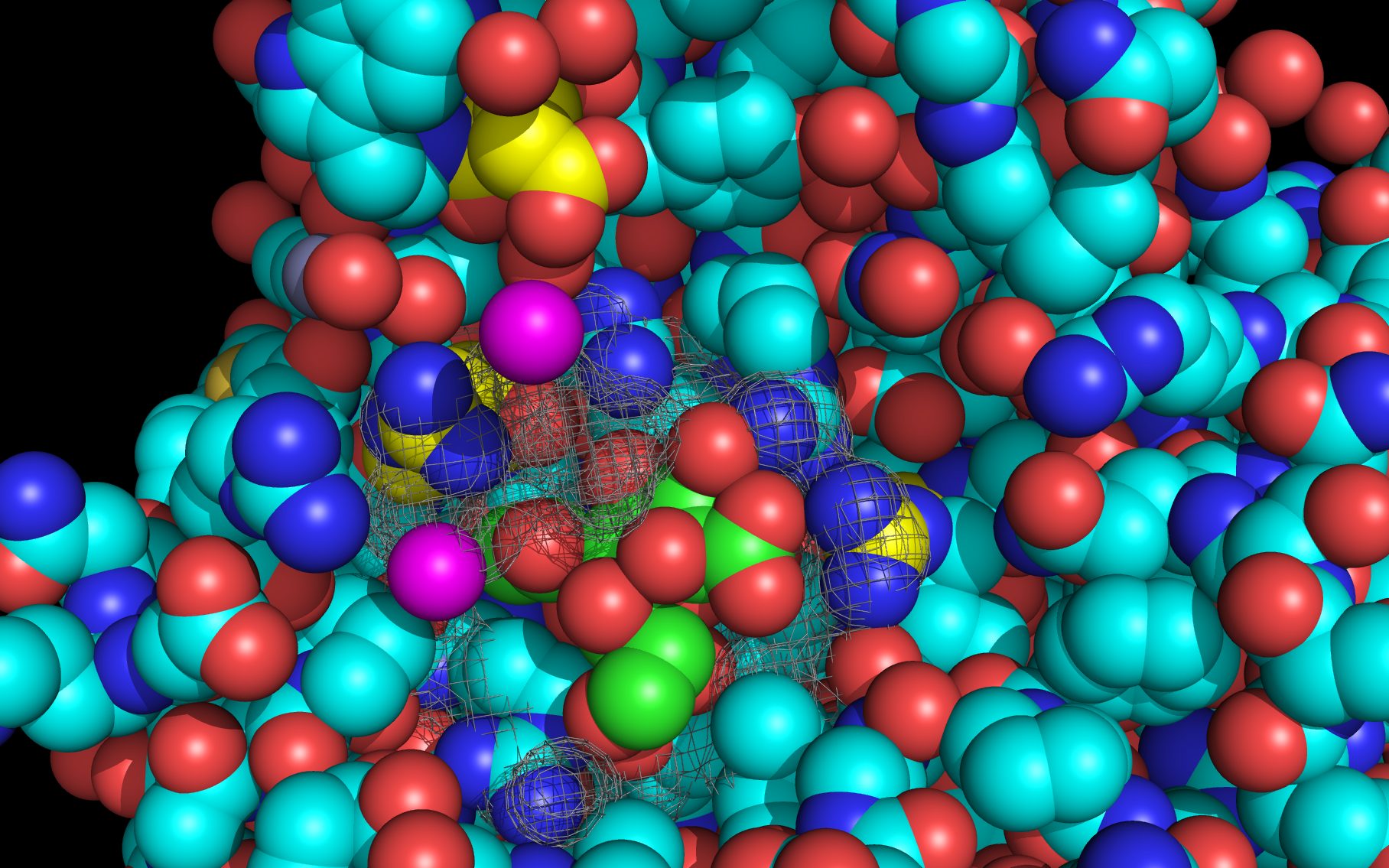
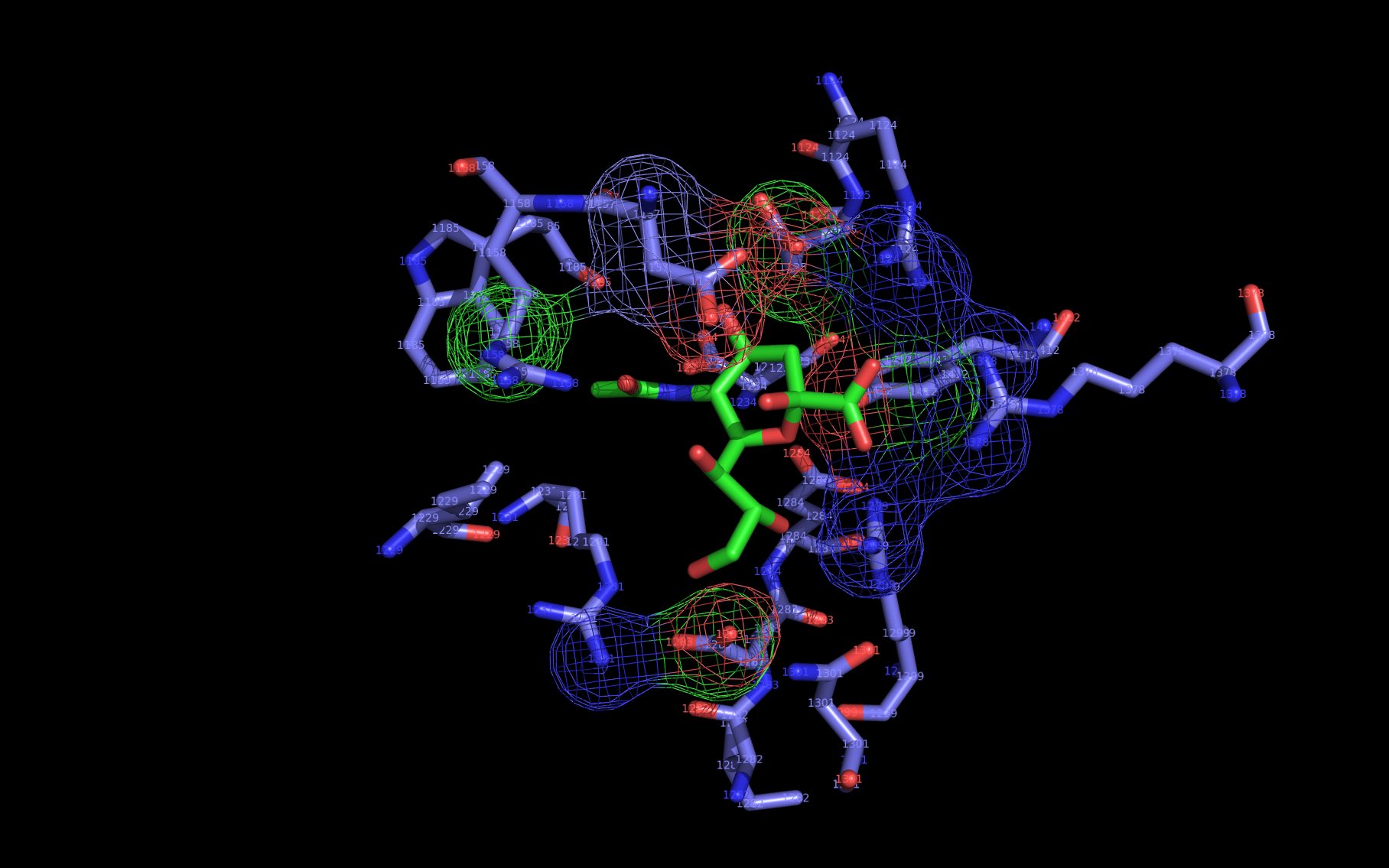
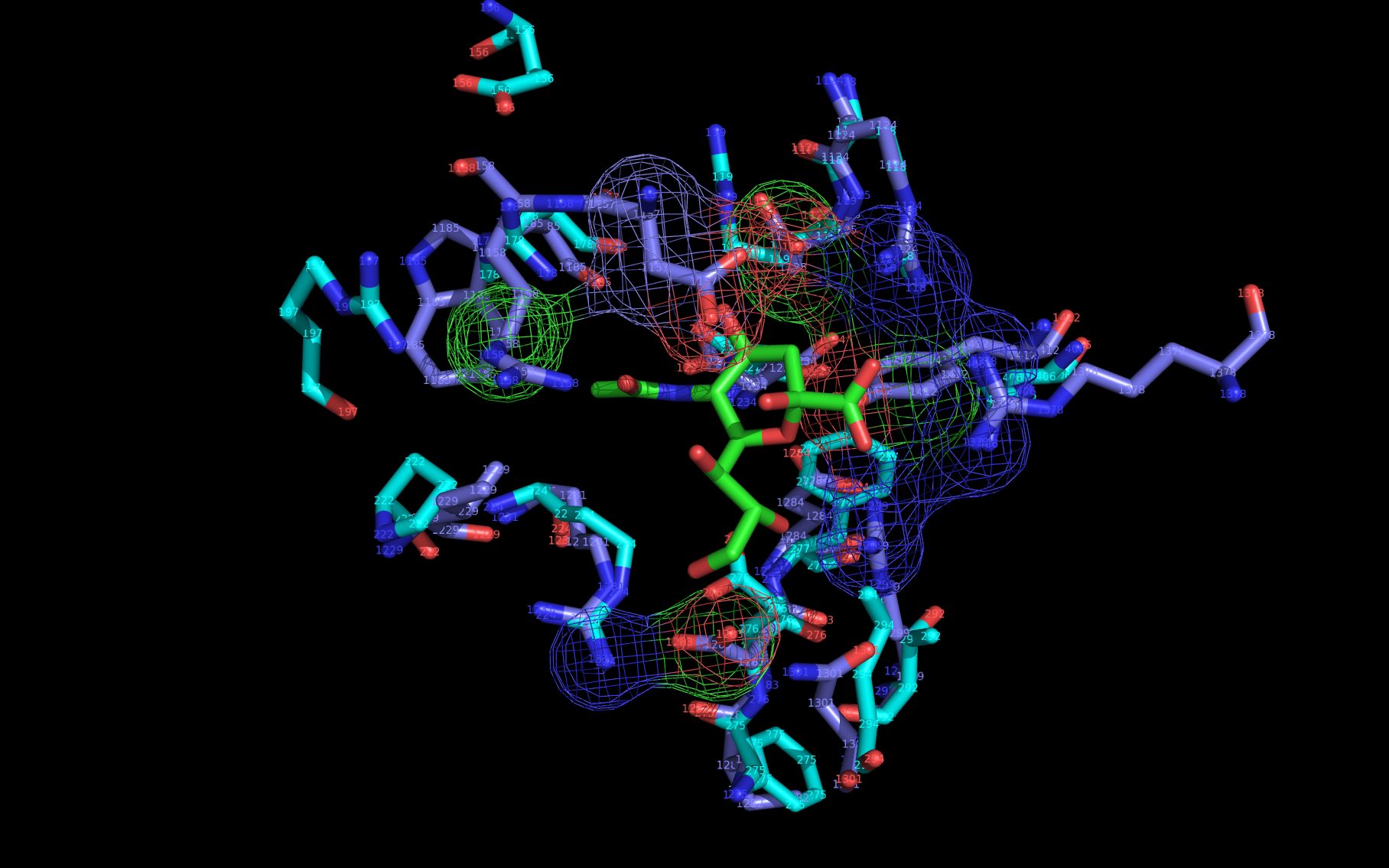
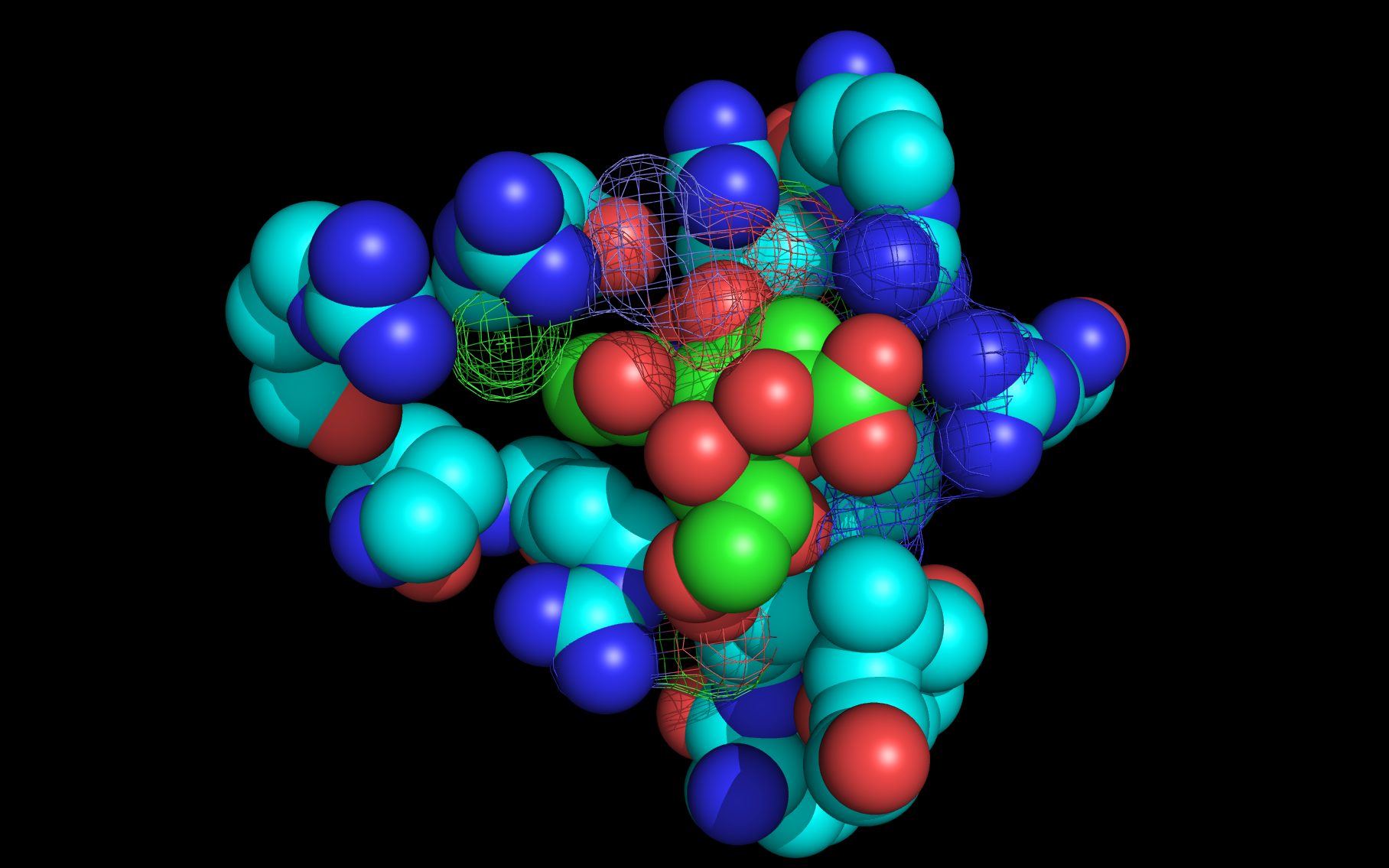
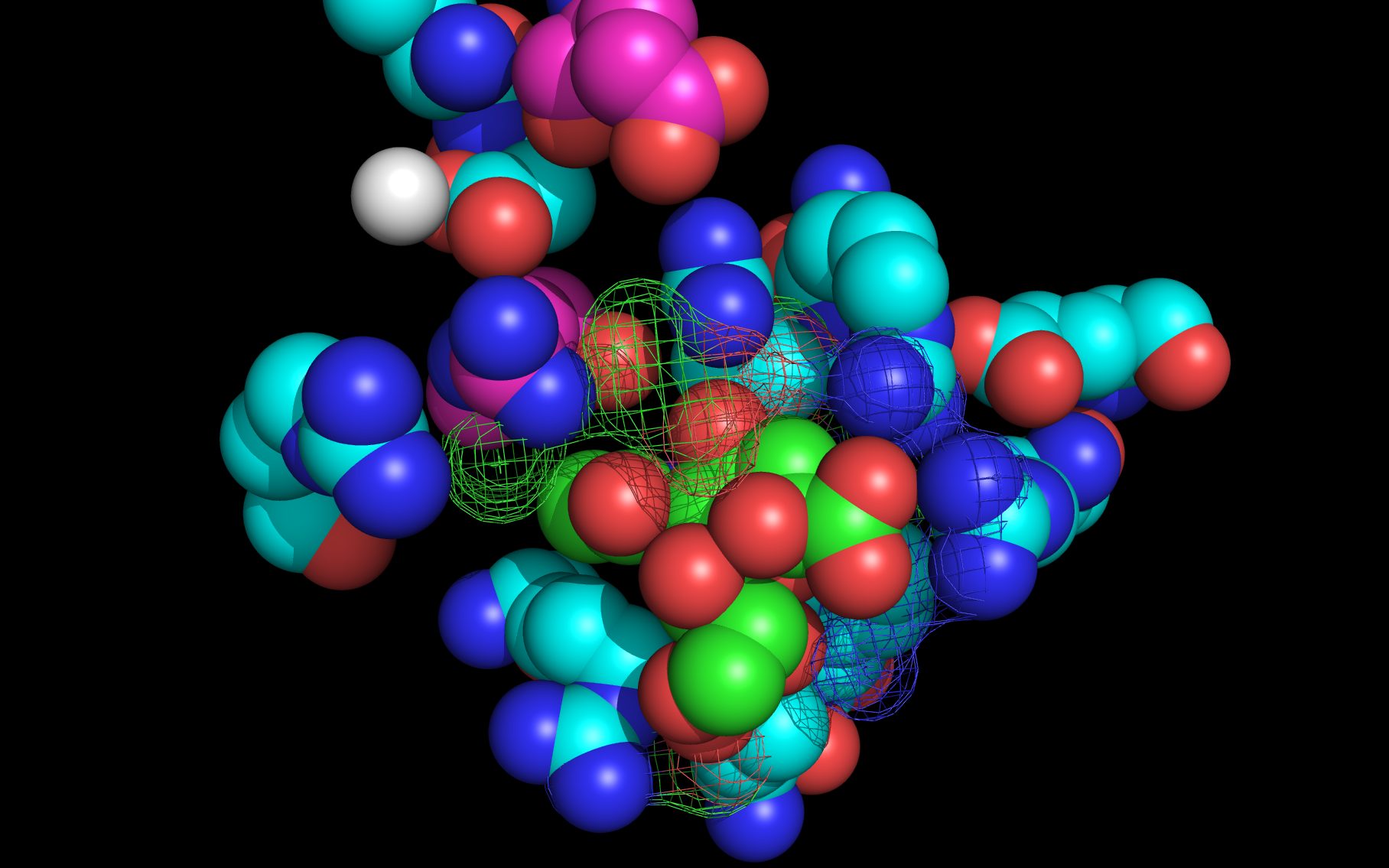
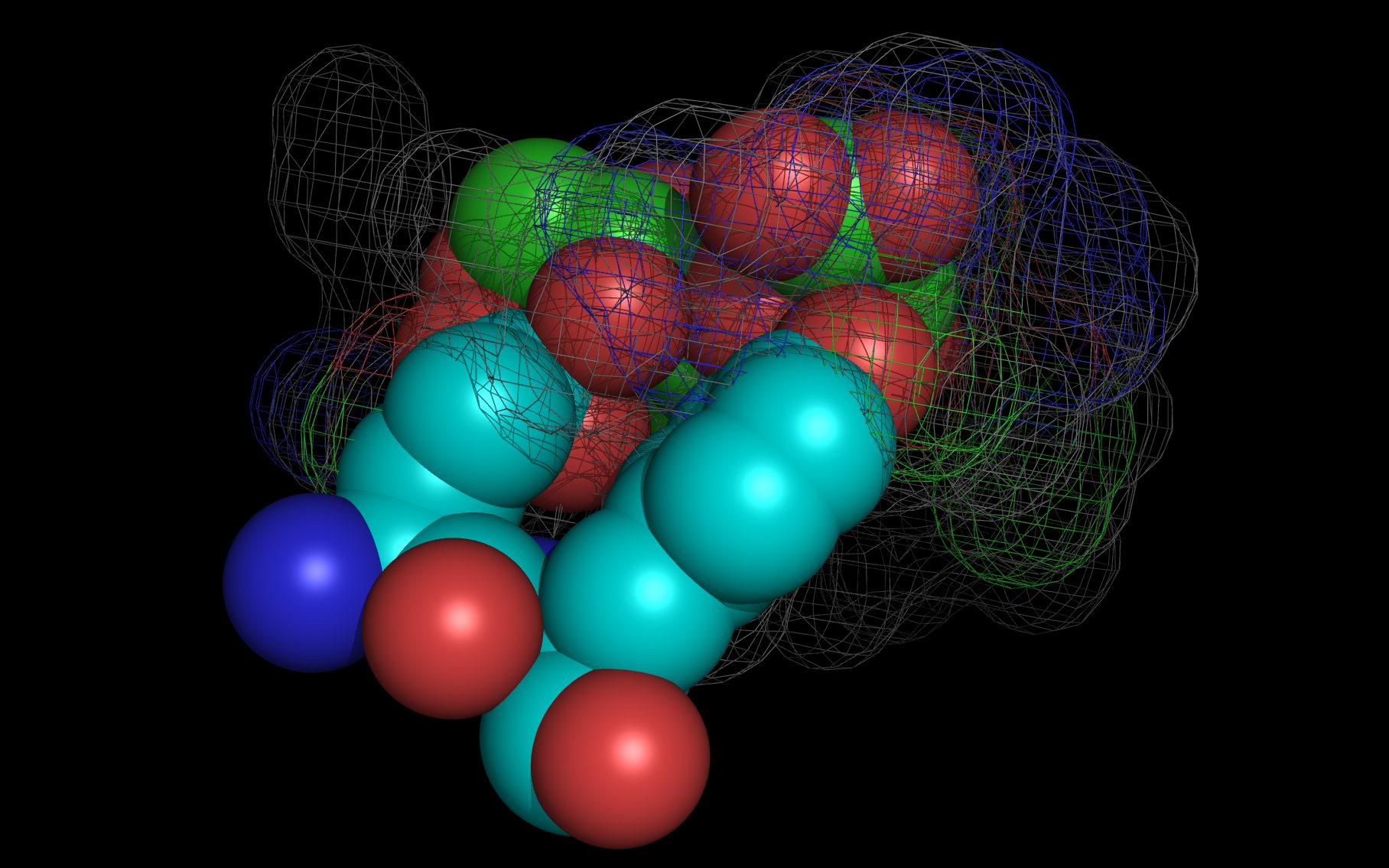
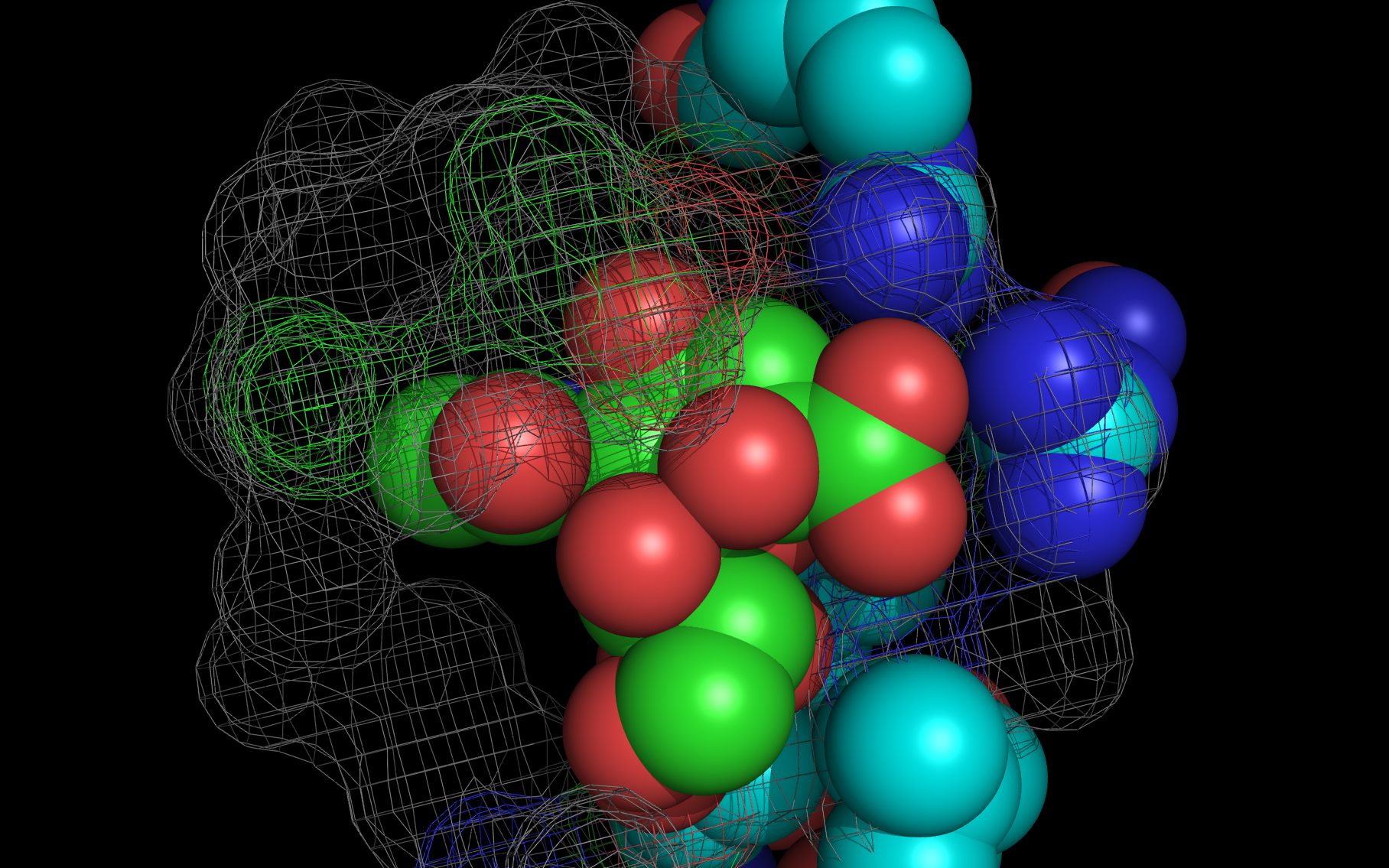
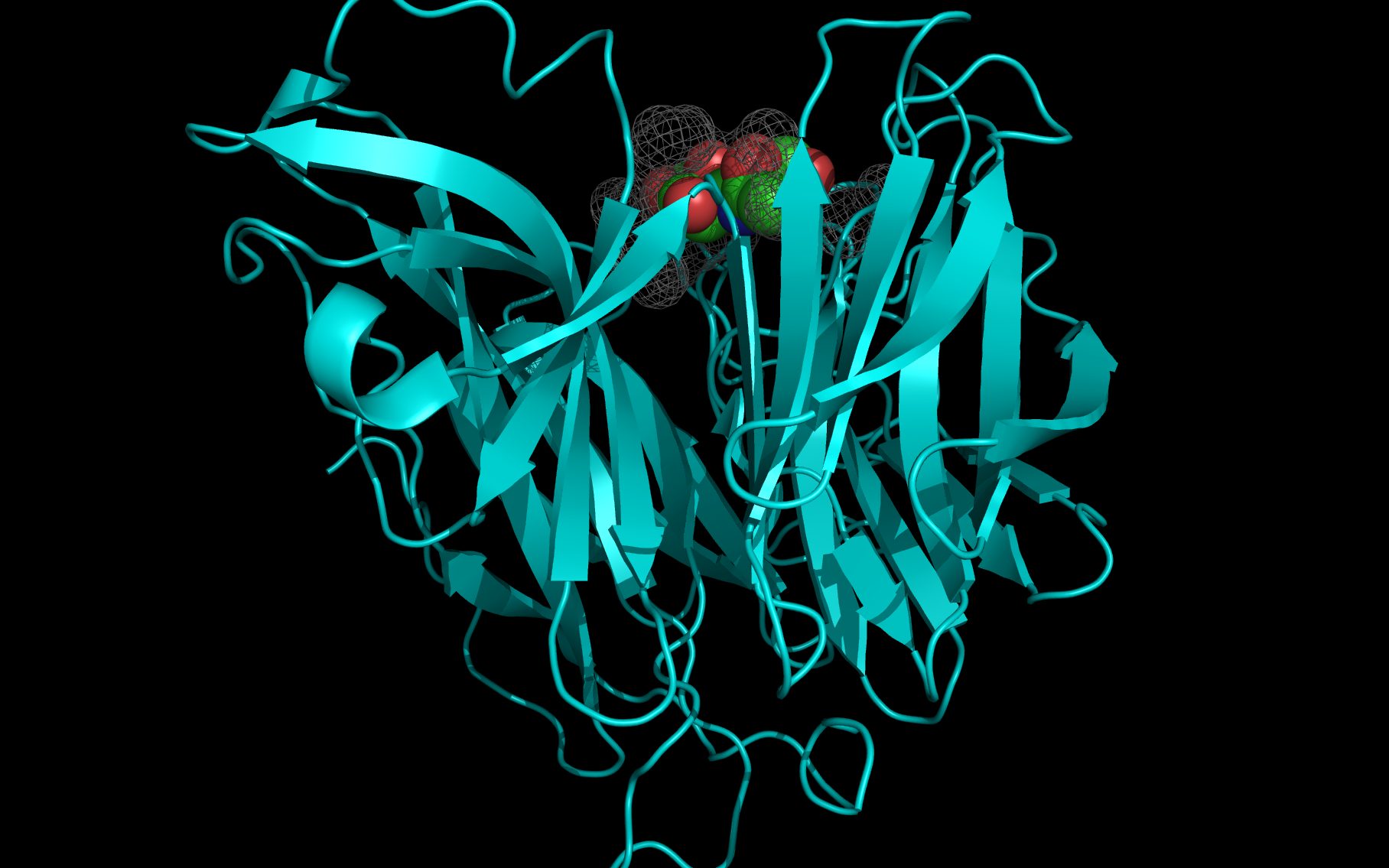
Shown here is sialic acid in the wwavePDB-identified putative sialic acid binding site of the bat viral influenza N10 neuraminidase. Publications6,7 of the structures (4VFK6 and 4GEZ7) of this bat viral influenza N10 neuraminidase have not identified this site and indicate that the N10 neuraminidase does not process MUNANA. wwavePDB analysis delivers a putative sialic acid binding site with functional features consistent with catalysis. This study suggests that the N10 neuraminidase may function under conditions different from the consensus neuraminidases. The wwavePDB-identified putative sialic acid binding site may not be able to bind and process MUNANA as the large size of MUNANA would prevent E153 to E156 loop movement.
wwavePDB produces “WWaveMarkers”, sets of atoms that are used to reorient structures relative to one another. See the discussion of WWaveMarkers and the atoms used for neuraminidase reorientation. WWaveMarkers were used to orient the N1, N6, N8, and N9 neuraminidases and associated bound compounds (sialic acid, oseltamivir, zanamivir, and peramivir) relative to one another. wwavePDB also identifies atoms in the promximity of bound compounds; the combined surfaces of these atoms are called “WWaveCores”.
WWaveCores were produced by wwavePDB from analysis of the N1, N6, N8, and N9 neuraminidases and their bound compounds. Sialic acid was oriented relative to the WWaveCores. The N10 structure was reoriented onto the N6 WWaveCores using WWaveMarkers. 1W1X1 and 4VFK6 were used as reference structures for identifying the components of the N10 neuraminidase putative sialic acid binding site. Selected side chain positions in 4VFK6 were altered in order to place wwavePDB-identified 4VFK6 atoms into WWaveCores. The sialic acid binding site was formed by reorienting wwavePDB-identified N10 neuraminidase side chain atoms to fill the consensus neuraminidase WWaveCores. The twwistPDB-reoriented positions of the residues of 4GEZ7 are substantially the same as 4VFK6 but the 4GEZ7 is not shown in the example images because its structure is so close to 4VFK6 as to add no value in the description.
Selected bound water oxygens are colored magenta .
wwavePDB-reoriented sialic acid spheres are color-coded according to element ( C N O ).
wwavePDB-reoriented mesh WWaveCores are colored grey .
wwavePDB-selected and reoriented 4VFK6 N10 neuraminidase residue spheres are color-coded according to element ( C N O ) with the exception that the E153, R178, and R406 residue atom spheres coordinating the sialic acid are colored ( C N O ).