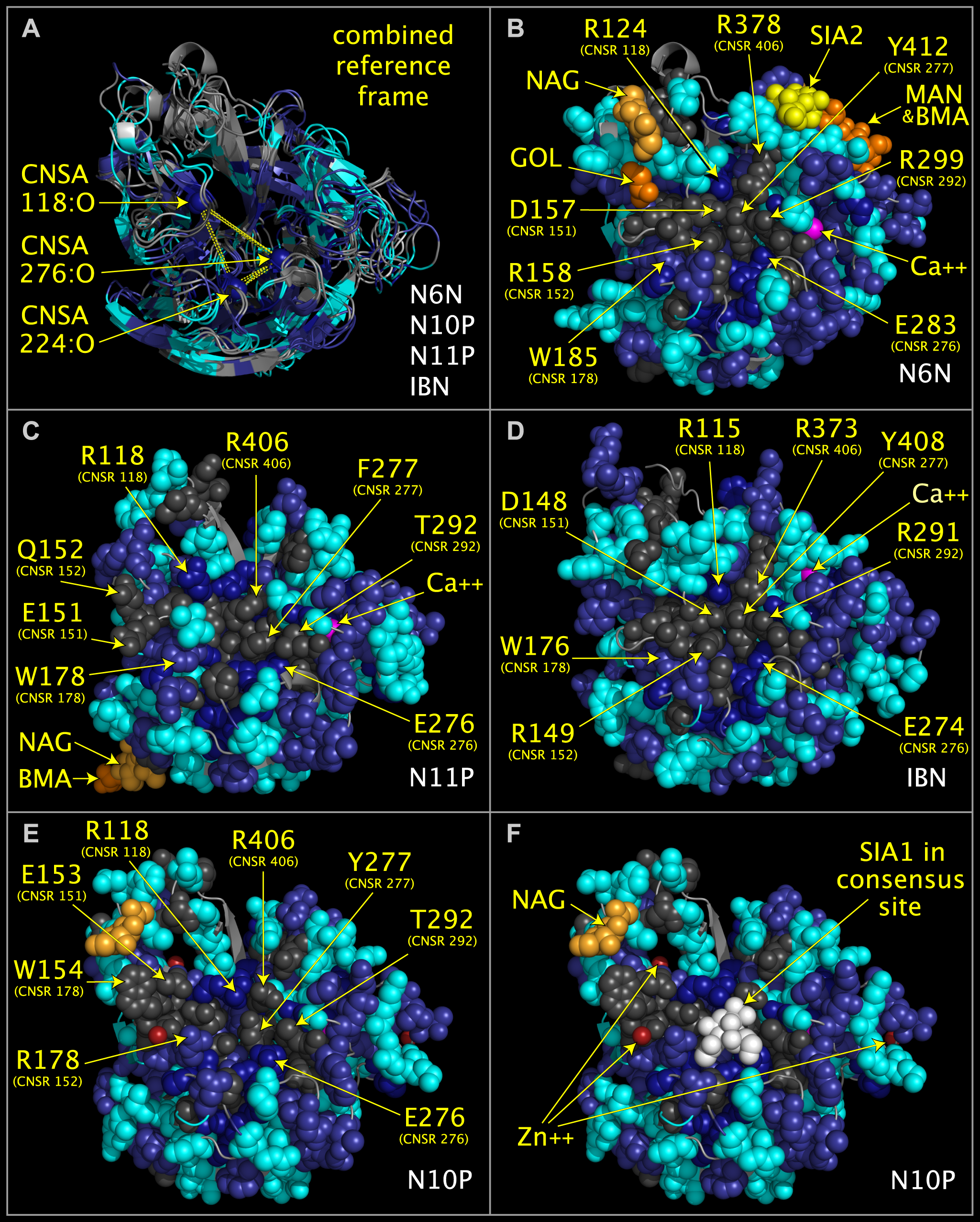
Figure 2. Consensus active site components of N6 neuraminidase (“N6N”), N10 protein (“N10P”), N11 protein (“N11P”), and influenza B neuraminidase (IBN).
Structure ribbon and residue spheres are color-coded at each structurally aligned position as in the ‘FIG2COL‘ row in Fig. 1. Fig. 2A shows structure ribbons representing influenza A N10P [1], N6N [2], N11P [3], and IBN [4]) structures superposed using CNSA118:O, CNSA224:O, and CNSA276:O atoms. Figs. 2B-2F show structure ribbons and residues spheres of N6N (Fig. 2B), N11P (Fig. 2C), IBN (Fig. 2D), and N10P (Figs. 2E-2F). Fig. 2F also shows sialic acid (white spheres) positioned relative to N10P by its superposition onto N6N, which was crystalized with sialic acid in its binding pocket. Fig. 2E and 2F residues side chains in the area of the sialic acid have been repositioned slightly from the crystal structure positions to approximate the positions of the corresponding superposed N6N residues but no other side chain or main chain atoms have been moved.